Your basket is currently empty!
Your basket is currently empty!
Our unique solutions for immunology research can save you time, money and give you clarity, whether your objective is basic research, preclinical or clinical development.
Our ClientsA new era of target-binding reagents beyond research antibodies: Ankyrons® offer affordable high-quality customized recombinant monoclonal binding reagents for any application and protein target. Find out more
Ankyron Catalog


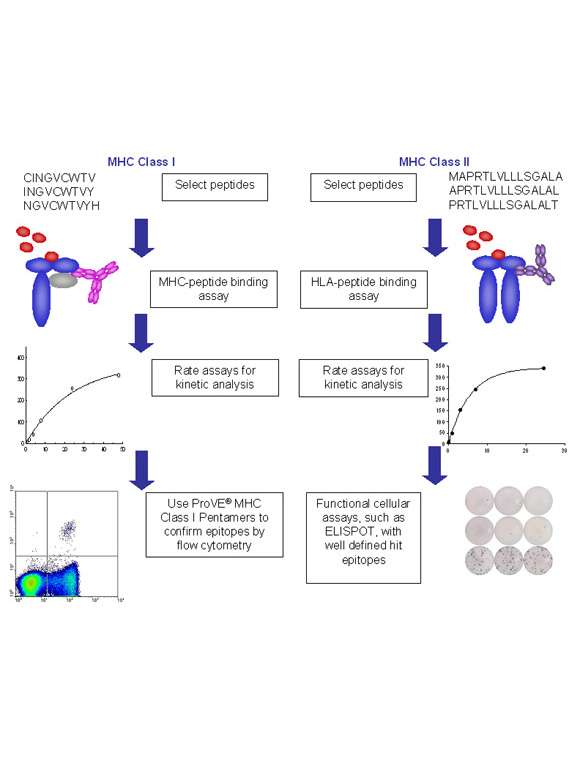
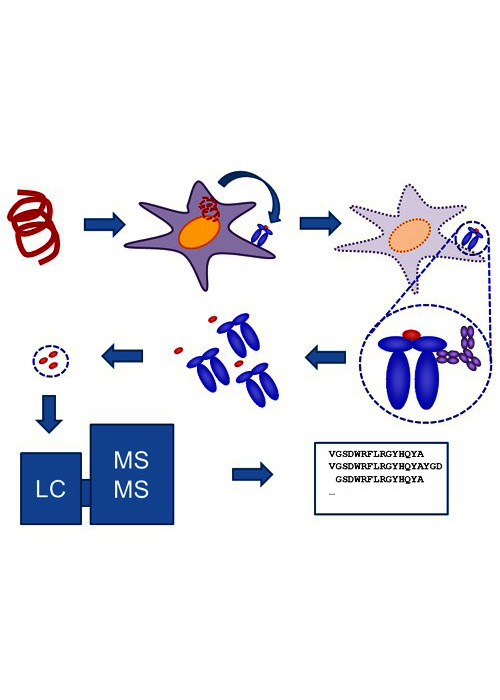

October 2024
A strategic partnership between The Pirbright Institute and ProImmune aims to develop target-binding reagents that significantly accelerate research in animal health.
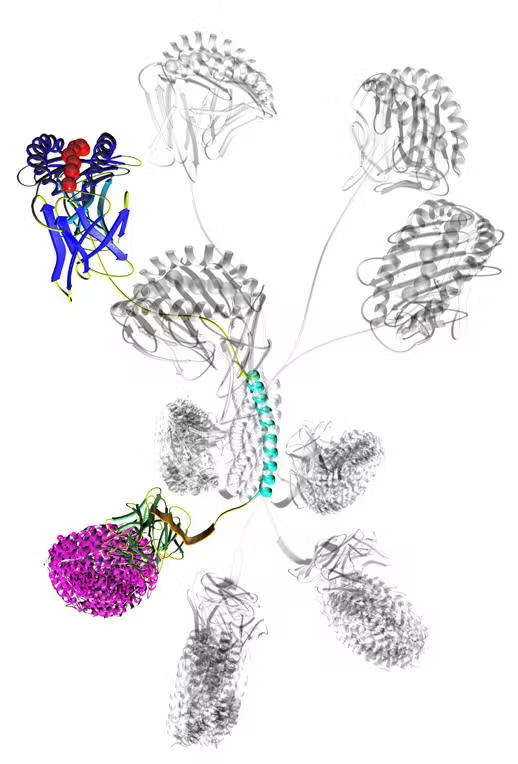
Ankyrons™ are small, recombinant, target binding ankyrin repeat proteins developed by ProImmune. They are directly selected in vitro from a trillion-clone library in ribosome display and, like antibodies, can bind with high affinity to almost any target.
Due to their superior properties compared to antibodies, Ankyrons™ open new avenues for scientists to study cell behaviour, offering an alternative to traditional research antibodies. The high specificity, sensitivity and reproducibility of Ankyrons™ means they can be broadly applied in biomedical science and can be targeted against all structural proteins as a cost-effective alternative to generating custom antibodies.
In this collaboration, ProImmune is aiming to establish new Ankyrons™ for a range of crucial veterinary research applications where antibodies were previously unavailable. ProImmune has already generated and provided new Ankyrons™ to Pirbright for further study and validation, including for a broad range of bovine, porcine, avian and mosquito protein targets, as well as targets for significant animal diseases including Foot and Mouth Disease and African Swine Fever – all areas that have been underserved by traditional antibody technology.
Pirbright’s Director of Research, Professor John Hammond, said: “The collaboration brings mutual benefits to address global animal health challenges. Multiple research groups across the Institute have worked with ProImmune to develop novel Ankyrons™ that address targets from across the tree of life including mammals, insects and viral proteins.”
Dr Linda Tan, Chief Scientific Officer, ProImmune, said: “Ankyron™ technology gives researchers a better handle on biology. In this collaboration we have already demonstrated that novel Ankyrons™ can be provided rapidly to dozens of targets in areas totally unaddressed by existing research tools. We believe that over time Ankyron™ technology will have a profound impact in the wider study of biology across species and the results from this collaboration are already testament to this.”
ProImmune and the Pirbright Institute hope to make significant strides in addressing global health challenges through the development of this cutting-edge research technology that will be available to researchers world-wide, studying animal physiology and disease.

January 2019
Mastering Immunity Europe 2019 will be taking place at Wolfson College, University of Oxford on 21 March. Speakers have now been announced and registration is now open.

September 2018
used by team at GSK to study mRNA vaccines that can protect against several Flu strains
Magini D. et al., PLOS One (2017)
Self-Amplifying
mRNA Vaccines Expressing Multiple Conserved Influenza Antigens
Confer Protection against Homologous and Heterosubtypic Viral
Challenge
http://dx.doi.org/10.1371/journal.pone.0161193
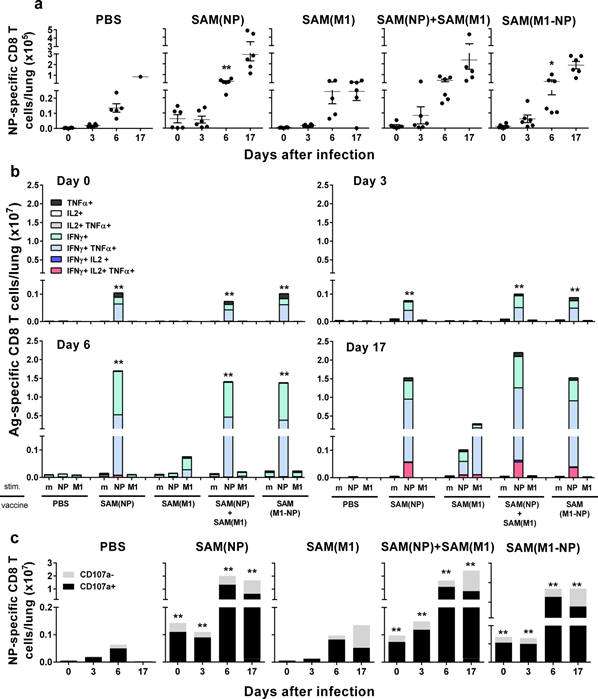
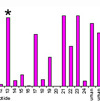
Figure:
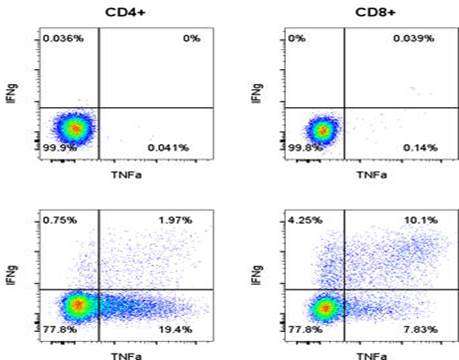
NP-specific
CD8+ T-cell responses in lungs after influenza
challenge. BALB/c mice
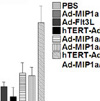
were immunized i.m. twice, 8 weeks apart, with PBS, 0.1 μg of
self-amplifying mRNA (SAM®) vectors SAM(NP), SAM(M1),
SAM(M1-NP), or with 0.2 μg of SAM(NP)+SAM(M1). Four weeks after
the second immunization, mice were infected with PR8 virus.
NP-specific CD8 T cells recruited in the lungs after the
infection were characterized by flow cytometry. (a) Numbers of
NP-specific CD8+ T cells determined using
Pro5® MHC Pentamers. Data are from individual mice (depicted as dots), while solid lanes
indicate the mean±SD. (b) Cumulative frequency of Ag-specific,
cytokine-secreting CD8+ T cells, indicated as
absolute number per lung. The color code represents the
different combinations of cytokine produced by the respective
cells after in vitro stimulation with medium (m), NP147-155
peptide (NP), or M1 peptide pool (M1), as indicated. (c)
Absolute number of NP-specific CD8+ T cells positive
(black bar) or not (grey bar) for CD107a. Data derived from two
independent and merged experiments. Statistical analyses were
performed using the Mann-Whitney U test. *p<0.05; **p<0.01
compared to the PBS-treated group.
http://dx.doi.org/10.1371/journal.pone.0161193.g006
Abbreviated abstract:
Current hemagglutinin (HA)-based seasonal influenza vaccines induce
vaccine strain-specific neutralizing antibodies that usually fail to
protect against mismatched circulating viruses. Inclusion of
conserved proteins such as the nucleoprotein (NP) and the matrix
protein 1 (M1) can increase effectiveness by eliciting
cross-reactive T-cells. However, efficient priming of T-cell
responses requires the right delivery system. Here we show novel
self-amplifying mRNA (SAM®) vectors expressing influenza
NP (SAM(NP)), M1 (SAM(M1)), and NP and M1 (SAM(M1-NP)) delivered
with lipid nanoparticles (LNP) induce robust polyfunctional CD4 T
helper 1 cells, while NP-containing SAM also induced cytotoxic CD8 T
cells. Robust expansions of central memory (TCM) and
effector memory (TEM) CD4 and CD8 T cells were also
measured. An enhanced recruitment of NP-specific cytotoxic CD8
T cells was observed in the lungs of SAM(NP)-immunized mice after
influenza infection that paralleled with reduced lung viral titers
and pathology, and increased survival after homologous and
heterosubtypic influenza challenge. We show for the first time that
the co-administration of RNA (SAM(M1-NP)) and protein (monovalent
inactivated influenza vaccine (MIIV)) was feasible, induced
simultaneously NP-, M1- and HA-specific T cells and HA-specific
neutralizing antibodies, and enhanced MIIV efficacy against a
heterologous challenge.
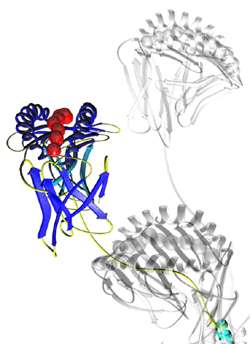
Pro5® MHC Pentamer

Pro5® MHC Class I Pentamers are the market leading product for detecting antigen-specific T cells, you can count on consistent performance backed up by unrivalled customer service.
To ensure uniformity and consistency, each Pentamer is purified four times before being subjected to rigorous quality control acceptance testing.

January 2016
Oxford, UK, 14 January 2016
ProImmune, a leader in services for understanding and managing adaptive
immune responses, announced today the introduction of ProT2™,
a range of Human MHC Class II tetramer reagents for tetramer analyses in
the study of antigen-specific CD4+ T cell immune responses. ProImmune’s
offering represents the broadest choice of ready to order catalog and
custom items in this field and will enable research into CD4+ T cell
responses in a variety of disease areas including cancer, infectious
diseases, allergy, autoimmunity and transplantation. Another key area
for this product range is the detailed study of T cell help in unwanted
immune responses to biologics.
CD4+ T cell
responses are at the heart of steering the adaptive immune system
directing both cytotoxic T cell responses and antibody responses
mediated by B cells. ProT2™ MHC Class II Tetramers allow the direct
detection and separation of single antigen-specific CD4+T cells
accurately by flow cytometry.
Nikolai Schwabe, CEO of ProImmune commented: “By introducing a broad and growing
ProT2™ Tetramer product range we intend to give MHC Class II tetramer
analysis the emphasis it deserves in the study of CD4+ T cell responses
which play such a pivotal role in adaptive immunity across so many
disease and treatment modalities.”
ProT2™ Class II MHC Monomer reagents are supported by ProImmune’s expert
customer service and application support, helping new users in
particular to establish and optimize robust protocols in their own
laboratories.
This addition to ProImmune’s class II reagent offering further complements an
already extensive range of services for evaluating CD4+ T cell
responses, including ProImmune REVEAL® MHC peptide binding assays, CD4+
T cell and DC T cell proliferation assays and standard immune monitoring
services through ELISPOT and flow cytometry based assays.
For more information on ProT2™ reagents and their availability, or to place an
order, please visit:
www.proimmune.com
email enquiries@proimmune.com, or call + 1 888 505 7765 in the United States or + 44 870 042 7279 in other countries.
-ENDS-
Notes to Editors:
About ProImmune –
www.proimmune.com
ProImmune’s mission is to
be a partner of choice for understanding and managing adaptive immune
responses. It does this by offering unique solutions for preclinical and
clinical immunology research, including a comprehensive and integrated
antigen characterization and immunogenicity testing platform, and
products and services for tracking antigen-specific immune responses
with state-of-the-art ELISpot and flow cytometry techniques. ProImmune
is committed to helping researcher’s achieve success through product
innovation, responsive service and focused application support, saving
time and money, and reducing risk.
For more information, please contact
At ProImmune:
Jeremy Fry,
Tel: +44 (0) 870 042 7279
Email: enquiries@proimmune.com
Media enquiries:
Sarah Jeffery
Zyme Communications Ltd
Tel: +44 (0) 7771 730 919
Email: sarah.jeffery@zymecommunications.com

11 Nov 2025 to 13 Nov 2025

07 Oct 2025 to 10 Oct 2025

17 Aug 2025 to 22 Aug 2025

11 Aug 2025 to 14 Aug 2025

16 Jun 2025 to 19 Jun 2025

20 May 2025 to 22 May 2025

12 May 2025 to 16 May 2025

07 May 2025 to 09 May 2025

05 May 2025 to 07 May 2025

03 May 2025 to 07 May 2025

01 Apr 2025 to 02 Apr 2025

30 Jan 2025 to 30 Jan 2025

Dr. Tim Hickling discusses the importance in the application of immunogenicity prediction and mitigation in the production of biotherapeutics. He outlines some of the current methods for predicting immunogenicity and highlights the need for better accuracy in predictions.

Professor Tao Dong from the University of Oxford discusses her latest work in memory T cell responses in convalescent SARS-CoV-2 patients in the UK, highlighting the information from her publication in Nature Immunology and the use of ProImmune Pro5® Pentamers in her work.

Dr. Amy Rosenberg of CDER, FDA discusses the clinical context for the reappraisal of immunogenicity of therapeutic proteins that are being used to treat COVID-19. She highlights the importance of understanding the autoinflammatory or autoimmune response following infection SARS-CoV-2 and its potential to increase immunogenicity to self proteins leading to autoimmune and autoinflammatory disorders. This indicates the high probability that this could lead to higher immunogenicity in patients to therapeutics sued to treat SARS-CoV-2.

Dr. Nina Le Bert from Duke NUS, discusses her recent involvement in the study of SARS-CoV-2 specific T cell immunity, specifically looking at the induction and persistence of T cells using an overlapping peptide library of specific proteins in SARS-CoV-2 to test convalescent patients' IFN-gamma responses to these peptides. Using a cohort of individuals who were infected and recovered in 2003 with SARS, and a similar approach of overlapping peptides from SARS-CoV, she is able to show there are still persistent ex vivo T cell responses. Importantly, she also highlights the cross reactivity of NP peptides from SARS-CoV and SARS-CoV-2.
CAR-T cells are an engineered T cell therapeutic in which a chimeric antigen receptor (CAR) (a foreign protein) is expressed on the surface of autologous T cells. This foreign CAR protein has the potential to elicit immune responses from the host immune system.
The immune response could be (a) humoral, resulting in the formation of Anti-CAR antibodies and/or (b) cellular, resulting in the development of cytotoxic T cells that are specific to the CAR-T cell. A host vs CAR-T response may result in (a) the neutralization of the CAR, rendering the CAR-T therapeutic ineffective (b) or result in the CAR-T cells being killed by a cytotoxic T cell response akin to a host vs graft response. This may manifest as lack of persistence of CAR-Ts and as loss of efficacy. There may be other reasons for loss of efficacy, and hence understanding if immunogenicity is the reason for loss of efficacy will be key in determining whether a second dose may be meaningful, the timing of this second dose, the impact of lymphodepletion on immunogenicity etc.

Katerina describes the process of codon optimization and the role that it plays in a protein's rate of translation, expression and conformational properties. She describes the presence of synonymous mutations in various diseases and the immunogenicity implications relating to those mutations. She uses Factor IX as a model to describe the workflow to generate variants using CoCoPUTs and the evaluation of those variants for protein expression, confirmational differences, peptide presentation and many more attributes.

Beatriz discusses the use of cancer vaccines to transition ‘cold’ tumors in patients to ‘hot’ tumors. The UPenn group used Next Gen Sequencing to catalog tumor mutations and generated autologous dendritic cell vaccines to evaluate the immunogenicity of missense mutations and define the nature of patient T cells specific for melanoma neoantigens. They found that that immunological ignorance of clonal neoantigens is the basis for ineffective T cell immunity to melanoma and that therapeutic vaccination as an adjunct to checkpoint inhibitor treatment, should be considered to increase the breadth and diversity of neoantigen-specific CD8+ T cells.

Antigen processing and presentation by HLA class II is a complex process and current in silico predictions are not able to account for all the factors involved in immunodominance/immunogenicity as they focus only the binding component of the process. Generation of large training datasets from single HLA class II allele cell-based or cell-free MAPPS-like platforms is enabling development of algorithms for identification of naturally processed ligands. The combination of binding and processing is a more reliable approach to the identification of viable CD4+ T cell epitopes, and to rank molecules. In silico prediction still needs to be part of a more complex immunognicity risk asessment strategy.

The immune response plays an important role in fighting cancer; however, the tumor environment is immunosuppressive and limits effective anti-tumor immunity. A new and promising strategy of tumor immunotherapy blocks pathways used by tumors to inhibit anti-tumor immunity. This inhibitory strategy is called checkpoint blockade. One key immunoinhibitory pathway that inhibits tumor specific immunity is the PD-1 co-inhibitory pathway. This pathway consists of the PD-1 receptor and its ligands PD-L1 (B7-H1) and PD-L2 (B7-DC). The PD-1 pathway plays critical roles in maintaining immune control, and is a key mediator of T cell dysfunction (“exhaustion”) in cancer and chronic infections. This pathway is a promising therapeutic target in cancer. The remarkable effects of PD-1 pathway blockade in cancer demonstrate the key role of this pathway in inhibiting anti-tumor immunity. However, there are multiple co-inhibitory pathways that that limit T cell function, and these have become targets for cancer therapy. This talk will discuss the multifaceted immunoregulatory roles of PD-1 and its ligands in controlling T cell activation, tolerance and exhaustion. The role of the PD-1 pathway in cancer as well as therapeutic strategies that combine PD-1 blockade with other therapies also will be discussed.

The gastrointestinal (GI) tract is home to trillions of commensal bacteria that play an important role in nutrition, immune system development and host defence. In inflammatory bowel disease (IBD), a chronic debilitating disease of the gastrointestinal tract, there is a breakdown in the healthy dialogue between our body and our microbial residents resulting in chronic immune attack in the bowel. In this presentation I will review key host and microbial pathways that maintain intestinal homeostasis and discuss how understanding these pathways may provide new therapies for the treatment of chronic inflammatory diseases.

Studies over the past 20 years have clearly defined and extensively characterized Foxp3+ T regulatory cells (Tregs) as major players in the control of most aspects of immune responses as well as playing a potential role in non-immunologic sites (fat, muscle). We are now at a point where studies have been/or will be initiated in the clinic using cellular biotherapy with Tregs, augmentation of Treg numbers/function with IL-2, and modulation of Treg function with small molecules and antibodies. Yet, many factors involved in Treg biology remain poorly studied. I will focus this talk on three key issues: 1) control of Treg homeostasis by TCR and co-stimulatory signals; 2) the role of transcription factors in modifying certain Treg functions; and 3) a critical review of suppressor mechanisms used by polyclonal and antigen-specific Tregs.

Dr. David Wraith, founder of Apitope, describes how peptide immunotherapies can induce antigen-specific desensitization. He describes how the REVEAL binding assay was used to identify pan-allele binding epitopes, which can servce as potential targets for tolerance induction.

Dr. Federico Mingozzi details the AAV-2 gene therapy model in Phase I/II trial hemophelia patients. He showed that a loss of human FIX expression was associated with expansion of capsid-specific CD8 T cells and demonstrated a dose-dependent T cell activity against the vector.

The immune response to protein-therapeutics (immunogenicity) is an important safety and efficacy concern during drug development and regulation. Non-clinical assays that can be used in the early stages of clinical development and to identify at-risk individuals and sub-populations in the clinic are an important unmet need. The so-called MHC Associated Peptide Proteomic (MAPPs) assay directly identifies peptides derived from a protein of interest on a donor’s MHC-II proteins. Here we have applied this technique to address several questions related to the use Factor VIII (FVIII) replacement therapy, in the treatment of hemophilia A. Over a dozen FVIII products are marketed but most fall into 3 categories: (i) Purified from human plasma (PD-FVIII). (ii) Full-length FVIII manufactured using recombinant DNA technology (FL-rFVIII). (iii) A truncated, beta-domain deleted rFVIII (BDD-rFVIII). We investigated whether there are differences in the FVIII peptides found on the MHC-II proteins of the same individual when incubated with products from the 3 classes. Our principal findings are as follows:
The number of unique FVIII peptides isolated, average length of peptides and range of peptide length were comparable for MHC proteins immunoprecipitated from cells from hemophilia A patients and healthy donors.
When cells from the same donor was exposed to FL-rFVIII in the presence and absence of PD-VWF, fewer peptides were recovered in the absence of PD-VWF; FVIII peptides not recovered in the presence of VWF map regions of VWF-FVIII interaction.
When dendritic cells from the same donor were incubated with FL-rFVIII or PD-FVII (both in the presence of PD-VWF); fewer FVIII peptides were recovered from the PD-FVIII. The peptides not recovered in the PD-FVIII did not map to the locations of FVIII-VWF interactions.
For each donor, FVIII peptides identified by the MAPPs assay exhibited affinities for that donor’s MHC variants which was orders of magnitude higher than the affinities of all overlapping peptides from FVIII and VWF, or when compared to a million random peptides obtained from the human proteome.
Zuben Sauna is a Principal Investigator and a Reviewer at the US Food and Drug Administration. His research interests lie in understanding the pharmacogenetic basis of the immune response to proteins used in therapeutic interventions as these affect efficacy and safety. His laboratory exploits a combination of computational, in vitro and ex vivo approaches to understand why some individuals and/or sub-populations develop immune responses while others do not. His work has published extensively in high impact journals such as Nature Biotechnology, Nature Medicine, Science, Science Translational Medicine and Nature Reviews Genetics. He received his Ph.D. from Poona University, India with subsequent training at the National Cancer Institute, Bethesda, USA.

Dr. Herman Waldmann reviews the mechanisms of tolerance induction and discusses strategies to evoke tolerance to immunogenic antibodies.

Accumulated evidence gathered in the last three decades demonstrated that some members of the Parvoviridae family, belonging in particular to the species Rodent protoparvovirus, have natural antineoplastic activity in cell culture and animal models, and that human tumor cells can be targets for this activity. In an immunocompetent rat glioma model, the H-1 parvovirus (H-1PV) was able to efficiently cure gliomas while raising an antitumor memory immune response. This oncosuppressive effect was observed after virus administration through intratumoral, intravenous and intranasal routes, and appeared to rely on both the direct oncolytic activity of H-1PV, and its handover to the host immune system. The parvovirus was also capable of inducing the regression of human glioma xenografts in immunodeficient rats. These studies have laid the foundations for the launch of a first phase I/IIa clinical trial in which H-1PV is currently undergoing evaluation for its safety and first signs of efficacy in patients with recurrent resectable glioblastoma multiforme progressing in spite of conventional therapies (ParvOryx01 trial). While no clinical conclusions, besides safety, can be drawn at this stage from the trial, there are intriguing laboratory measurements in resected tumors and patients’ blood. Immunohistochemical and FISH analyses provided evidence for virus efficiency in crossing the blood-brain barrier, spreading throughout the tumor and getting expressed in tumor tissues. The possible impact of these direct viral effects on the tumor fate has to be balanced against evidence of immune infiltration and vascular disruption in infected tumors, and of systemic immune responses in treated patients.

Bernard Maillère is currently director of research and head of the laboratory of immunochemistry of the cellular immune response at Commissariat à l'Energie Atomique (CEA), a French Governmental Research organism. His works mainly deal with the prediction of immunogenicity and identification of T cell epitopes in humans. Bernard is currently workpackage leader of the European IMI project ABIRISK dedicated to immunogenicity of therapeutic proteins


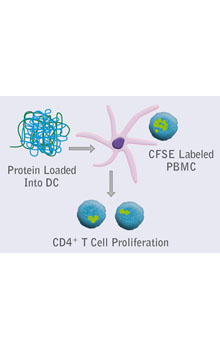
Integrated platforms can be used to mitigate immunogenicity risk and characterize immune responses during the drug design and development stages. ProImmune offers mutational activity mapping for optimal protein design, DC-T/T cell proliferation assays for biologic lead selection/optimization, a Mass Spectrometry assay for characterization of antigen presentation; HLA-peptide binding assays to characterize individual epitopes & undiluted whole blood cytokine storm assays.

Dr. Butler is Assistant Professor at the Princess Margaret Cancer Centre Tumor Immunotherapy Programme where is he is the Clinical Head of the Immune Monitoring Team. Before joining he was an Instructor in Medicine at Harvard Medical School and Clinical Fellow in Medicine at the Dana Farber Cancer Institute. Dr. Butler’s expertise includes clinical trials in a variety of immune therapies including adoptive T cell therapy and checkpoint blockade.

Immunogenicity of biologics in the clinic has the potential to cause severer sequela and alter or neutralize efficacy resulting in exposing patients to risk and terminated development programs. Pre-clinical risk assessment utilizing orthogonal methods has made it possible to prioritize drug candidates with a decreased immunogenicity risk or utilize protein engineering o reduce immunogenicity risk. Here we will present the immunogenicity tool box utilized at Bristol Myers Squibb to pre-clinically minimize immunogenicity risk.
Jochem Gokemeijer has been working at Bristol Myers Squibb for 15 years in Immunogenicity and biologics drug discovery. Before that he was involved in two alternative scaffold biotech companies Phylos and Adnexus Therapeutics. Jochem did his undergraduate work in the Netherlands and graduate work at the Dana Farber Cancer Institute.

Dr. Paul Moss describes the CD8 and CD4 T cell immune responses to CMV infection. He describes the importance of Class II Tetramers to better understand the interplay of a chronic infection and immune control.

Dr. Emilee Knowlton of ProImmune describes the tools for epitope discovery and immune monitoring, providing case studies and considerations for both pre-clinical and clinical

Competition for limited, cell extrinsic survival factors is a general feature of the peripheral selection checkpoints involved in B lymphocyte maturation, activation and memory. Perhaps the best-characterized example involves BLyS family cytokines and receptors, which governs survival and differentiation within B cell subsets. Discovery of the BLyS cytokine and receptor family has proven a watershed event, significantly advancing our understanding of B lymphocyte selection and homeostasis. This family includes two ligands, BLyS (also termed BAFF) and APRIL; as well as three receptors, BR3 (also termed BAFFr), TACI, and BCMA. Members of this molecular family play critical roles in maintaining immunological tolerance, by impacting selection and survival in nearly all B cell subsets. Cells in the transitional and mature, pre-immune B lineage subsets rely on BLyS signals via the BR3 receptor for survival. In contrast to tolerogenic elimination at the BM immature B cell stage, the transitional checkpoint displays plasticity by integrating BCR-mediated selection with BLyS-mediated peripheral B cell homeostasis. Thus, when BLyS levels are elevated, transitional selection is “relaxed,” so that B cells that would normally be negatively selected instead survive to join mature naïve pools. Mounting, evidence suggests analogous competitive checkpoints for both germinal center B cells and plasma cells. In contrast to pre-immune pools, B cells in recently activated and antigen-experienced subsets shift their BLyS receptor profiles and hence their ligand reliance. For example, short-lived antibody-forming cells adopt a TACI dominated BLyS receptor signature, whereas germinal center (GC) B cells profoundly down-regulate TACI but retain BR3. The lack of TACI on GC B cells leads to a paucity of retained BLyS in the GC, such that the sole local source of BLyS in the GC is the TFH cell. Moreover, BLyS expression by GC TFH is crucial for appropriate GC evolution, since efficient affinity maturation fails if T cells are BLyS deficient. Finally, long-lived plasma cell pools express BCMA, shifting reliance to APRIL. Considered together, these observations suggest that deliberately altering BLyS levels might be a means for manipulating B cell repertoire selection in order to (1) restore self-tolerance in autoimmunity, (2) remodel the repertoire to accommodate neo-self antigens introduced through biologicals, transplantation and gene therapy, or (3) temporarily expand B cell repertoire diversity to reveal novel, therapeutically useful specificities.

The central role of T cell responses to clinical successes in immuno-oncology has been well appreciated, yet few technologies have been designed to exclusively generate effective and sustained T cell responses in vivo. The DPX platform is a unique oil based formulation that entraps its components and facilitates active uptake by antigen presenting cells at the site of injection. This unique presentation to the immune system results in robust and sustained T cell responses. The lead clinical product, DPX-Survivac, employs the DPX platform to induce T cells targeting the shared tumor associated antigen survivin, which is overexpressed in many human malignancies. DPX-Survivac has been demonstrated to induce high levels of circulating, polyfunctional T cell responses to survivin in clinical studies of advanced ovarian cancer patients, and some of these responses have been associated with an ability to mediate tumour regression. Preclinical and clinical data have supported combining this T cell targeting treatment with other immuno-oncology agents, such as checkpoint inhibitors.

Dr Lea Bartsch from Massachusetts General Hospital discusses her work with HBV, in trying to better the understanding of the immunological alterations in order to improve treatment strategies for HBV infection. She highlights the use of ProImmune’s custom ProT2 Class II Tetramers in tracking HBV antigen specific CD4 T cells to validate new HBV epitopes and analyze the different CD4 T cell responses in chronic vs. acute infections.

Dr. Kellie Smith from Johns Hopkins, discusses her group's global study of the associations of neoantigen specific T cell responses with clinical outcomes to checkpoint blockades in cancer. The talk outlines the use of their MANAFEST T cell receptor sequencing technology, and by sequencing the CDR3 region, they are able to track clones that recognise the same antigen of interest. Showing that with lower clonality there is an association with higher residual tumour burden in patients. Pro5 Pentamers are also shown being used as an accurate way to validate their findings, by using them to track the neoantigen specific CD8 T cells. She also highlights the use of single cell RNA-seq in determining the phenotype of the clones in responding and non-responding patients.

Professor David Withers from the University of Birmingham discusses his recent work in tracking dynamic changes of lymphocytes within tumours, to understand and characterise the phenotype of cells entering, leaving and being retained in the tumour through the use of the transgenic Kaede Photoconvertible mouse model. He also discusses the use of Single Cell RNA-seq to capture the heterogeneity within the tumour infiltrating lymphocyte population, and describing the use of ProImmune Pro5® Pentamers to track tumour neo-antigen specific CD8 T cells.

Hilario Ramos, Molecular Templates, discusses the use of Egineer Toxin Bodies (EBTs) – molecules with an antibody domain for targeting that is genetically fused to a de-immunized Shiga-like toxin A subunit which drives apoptotic cell death. These molecules can drive cell death by apoptosis, and Hilario discusses novel uses for EBTs of delivering antigenic peptides to tumours, in a novel way to make the tumour cells visible to the immune system. Hilario highlights the use of ProImmune's Pro5® MHC Class I Pentamers in tracking Antigen Specific CMV CD8 T cells in order to help build their CTL models.

Dr. Yeojun Yun discusses Synteka Bio's NEOscan™, a neoantigen prediction algorithm used to improve the selection and prioritization of neoantigens for inclusion in cancer therapies.

Christine Falk graduated in Biology at the Ludwig-Maximilians-University in Munich and performed her PhD thesis at the Institute for Immunology at LMU Munich (director G. Riethmüller) in the laboratory of D.J. Schendel on autoimmunity and the regulation of NK-like T cells and NK cells. As a postdoctoral fellow at the new Institute of Molecular Immunology (director D.J. Schendel) at the Helmholtz Center for Environment and Health (GMGU), she studied recognition of virus-infected cells and tumor cells by T and NK cells. As senior postdoctoral fellow and head of the research group “Immune Monitoring” at the German Cancer Research Center DKFZ in Heidelberg from 2006 until 2010, she was working on the microenviroment of solid tumors and its impact on tumor recognition by T and NK cells. In her current position since 2010, she is head of the Institute of Transplant Immunology, IFB-Tx, at Hannover Medical School MHH where she has established an immune monitoring laboratory investigating adaptive and innate immune responses in the context of stem cell as well as solid organ transplantation, in particular lung, heart, liver and kidney transplantation. The addition of transplant immunology to her former field of tumor immunology is reflected by focusing on common immunological mechanisms for rejection of both tumors and allografts that may open new perspectives for immunological interventions to achieve tumor rejection on one hand and on the other hand, immunological tolerance, the ultimate goal of solid organ transplantation

In cancer cells, the disruption of normal biochemical signaling results in aberrant post-translational modifications, including protein phosphorylation. These mis-phosphorylated proteins can be processed into antigenic peptides by the cellular machinery, resulting in presentation of this novel type of antigen on MHC molecules. PhosphoSynVax is Agenus’s third heat-shock protein-based vaccine platform. PhosphoSynVax is designed to induce immunity against this novel class of phospho-neo-epitopes specific to malignant cells of multiple donors in various cancer types. Based on mass-spectrometry and bio-informatic approaches, we have a library of proprietary phosphopeptide tumor target antigens from different cancers, including lung cancer, specific leukemias, ovarian cancer, colon cancer and others. This approach may enhance immune system recognition of phospho-neo-epitopes, leading to the destruction of cancer cells in various patients across cancer types.

The ability to clear many intracellular infections is likely to require a consorted immune response that includes CD8+ T cells. Although a large number of approaches have been taken to generating such responses in humans, achieving the magnitude and functionality of natural viral infections has been difficult. The most promising approach published to date is to employ a heterologous prime-boost, usually using two replicating or non-replicating viral vectors. The Jenner Institute has extensively used a replication-deficient adenovirus followed by a replication-deficient pox virus, and achieve ELISPOT levels of balanced CD4+ and CD8+ T cells in the many thousands of spot forming units per million PBMCs. Vaccitech has in-licensed programs based on the use of this strategy to control chronic HBV and chronic HPV. The progress of these programs will be discussed.

Jonathan Silk is the Head of Cell Research at Adaptimmune (www.adaptimmune.com), a biotechnology company located in Abingdon and Stevenage, UK, and Philadelphia, PA, USA. Adaptimmune is a world leader in the TCR T-cell therapy space, utilizing engineered Specific Enhanced Affinity T-cell receptor-expressing T-cells (SPEAR T-cells) to target both solid and hematologic cancer types. Jonathan and his team are investigating Next Generation SPEAR T-cells, to improve the function of T-cells for adoptive T-cell therapy, including overcoming immune-resistance mechanisms. Jonathan received his BSc in Biochemistry from the University of Kent at Canterbury (UK), with an industrial placement at Merck Sharp and Dohme (Harlow, UK). He obtained a PhD in Immunology studying at the MRC Clinical Sciences Centre (Imperial College London, UK). Subsequently he worked with Professor Vincenzo Cerundolo at the University of Oxford, UK, as a Post-doctoral Scientist, developing a number of cancer immunotherapy research programmes including the use of ligands for invariant NKT-cells as adjuvants and investigating mechanisms of tumour escape.

Rodd Polsky is an Investigator responsible for immunogenicity assay development within the Bioanalysis, Immunogenicity and Biomarkers organization at GlaxoSmithKline. Rodd received his Bachelor’s degree in Biology from Drexel University. Since that time, Rodd has spent the past 25 years providing preclinical and clinical immunogenicity support, including assay and study design, development, validation, and study support for the detection of anti-drug antibodies against therapeutic proteins.

Dr. Aaron Mansfield of Mayo Clinic Cancer Center discusses the current immunotherapy treatments for patients with mesothelioma, the relatively low tumour mutation burden in those patients and alternatives to non-synonymous point mutations as sources of neo-anitgens. The use of ProImmune REVEAL technology allowed him to compare the MHC binding scores of selected peptides from chromosomal rearrangements, of usually non-coding DNA to discover if any of these peptides were neo-antigenic.

Karen discusses how to measure antigenic diversity of B and T cell immune responses and how the structural components of those antigens drive immunogenicity. She describes technologies such as custom protein-peptide microarrays to detect antibodies; a mass spec based program called MHC TreASUre Hunt for immune peptidome discovery; and Molecular Dynamic Modeling to model peptide MHC interactions based on existing crystal structures to help develop computational systems to predict TCR.

Vibha discusses a proactive risk based approach to assessing immunogenicity in early development. She describes a suite of in silico risk assessment tools in combination with in vitro methods to evaluate immunogenicity or product attributes and how, when used in combination, those methods can reduce false positive identification. The stages of when to implement and report the data from the risk assessment tools is described as well as the current challenges the FDA reviewers face when evaluating the data package.

Rao describes Multiple Myeloma, a plasma cell cancer and the biomarkers that can be used to classify and monitor the immune response through the progression of the disease. They Harvard group looked at myeloid derived suppressor cells, plasmacytoid DC, T-regulatory cells, TH17 cells and B cell subpopulations in the interrogation of the cancer. There was a humoral immunity deficiency, increased PD1 expression on effector cells and increased levels of PDL1 expression detected in the multiple myeloma patients.

Paul discusses the challenges facing the identification and diagnosis of Lyme disease. He describes the life cycle stages of the carrier, Borrelia burgdorferi sensu lato species, as well as the incidence and transmission of Lyme disease in the United States. There are differential expressions of antigenic surface proteins that occur due to bacterial transcriptional changes and throughout the course of the disease. Highly reactive serum was screened for responses against 21 different proteins using 80 overlapping peptides in the ProArray Ultra linear B cell epitope mapping tool to improve the specificity and accuracy of current diagnostic methods.

Patrick describes the Children’s National cellular therapy program for treating viral infections following transplant surgeries. He discussed the generation of donor-derived virus specific T cells from cord using either an adenoviral vector or peptide mixes and the recognition of atypical epitopes by these cells. The team has treated over 80 patients with virus specific T cells with between 70-80% response rate.

For forty years, I’ve been researching therapeutic antibodies with a particular interest in effector functions of the Fc region. With Herman Waldmann and Greg Winter, I took several of the early engineered antibodies into the clinic. We realised that activation of immune effector systems is not always desirable, but found that mutations designed to reduce binding to Fc receptors (e.g. ‘LALA’, aglycosyl) were not completely effective. At mAbsolve we have screened hundreds of new variants and identified some which are completely devoid of binding to Fc receptors.
Receptor binding is one thing, but cellular activity is more important. The modest binding levels of previous variants translated to quite substantial cellular activity, but our new variants remained completely negative. The risk of unwanted immunogenicity is always a concern when proteins are modified, but the variants gave no increased risk signals either in silico or in vitro.
The optimised Fc variant is comparable with wild type IgG for binding to FcRn, stability and manufacturability and is available for licensing.

Katie Ewer is Associate Professor and Senior Immunologist at the Jenner Institute, University of Oxford where she leads a program of exploratory immunology studying vaccine-induced immunity to malaria and outbreak pathogens, such as Ebola, MERS and now SARS-CoV-2. A key aim of her research is focused on characterizing T cell responses induced by vaccination with viral vectors. During the COVID-19 pandemic, she played a pivotal role in the clinical trials in an effort to find a vaccine, which has led to the successful roll-out of the first approved SARS-CoV-2 vaccine, ChAdOx1 in partnership with AstraZeneca in December 2020. In this talk she explains how the vaccine was developed in record-breaking time, the characterisation of the immune responses to the vaccine and the forthcoming challenges still faced including the question of booster doses, manufacturing and supply and how we respond to the next inevitable pandemic.

In this talk we learn how quantity and early induction of virus-sepcfici T cells is directly associated with mild disease; how higher functionality of SARS-CoV-2 specific T cells is found in assymptomatic cases of SARS-CoV-2 infection and how how early detection of SARS-CoV-2 specific T cells correlates with the the onset of vaccine efficacy.
Nina Le Bert is currently a Senior Research Fellow in the Emerging Infectious Diseases Programme of DUKE-NUS Medical School in Singapore. She earned her Master of Science degree at the Free University of Berlin in Germany, and her PhD from the University College London UK in 2011, working in the field of human innate and adaptive immunity. In 2014, she joined Professor Antonio Bertoletti’s lab, where she is investigating the virus-specific B and T cell responses during Hepatitis B Virus (HBV) infection to better understand their role in protection and/or liver pathology. Since the outbreak of the COVID19 pandemic, she is involved in studies of SARS-CoV-2 specific T cells.

In this talk we learn that SARS-CoV-2-specific PD-1+CD8+ T cells are not exhausted, but functional; that they are sustained for at least 8 months, as well as learning about the development of SARS-CoV-2-specific stem cell-like memory T cells (TSCM).
Eui-Cheol Shin received his M.D. (1996) and Ph.D. (2001) from Yonsei University, Seoul, Korea, and his postdoctoral training from National Institutes of Health, Bethesda, Maryland, USA. Then he joined Graduate School of Medical Science and Engineering, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Korea in 2007, where he is currently a professor. His laboratory, the Laboratory of Immunology and Infectious Diseases, focuses on the research of T cell responses in human viral disease and cancer.

In this presentation, Dr. Sette provides evidence that there are robust CD4+ and CD8+ T cell responses detected in uncomplicated SARS-CoV-2 convalescent cases; that reactivity is reproducibly detected in non-exposed subjects; specific CD4 and CD8 T cells targets in COVID-19 patients were identified; in acute and severe infection it appears that the speed of the adaptive response is key to protective immunity; that T cell responses are durable over at least 8 months and that there is negligible impact of SARS-CoV-2 variants on T cell responses.
Alessandro Sette has devoted more than 35 years in biotech and academia to understanding and measuring immune responses, and developing disease intervention strategies against cancer, autoimmunity, allergy, and infectious diseases. Dr. Sette’s laboratory is the world leader in the study of the specific structures, called epitopes, that the immune system recognizes. Dr. Sette has overseen the design and curation efforts of the national Immune Epitope Database (IEDB), a freely available, widely used bioinformatics resource. The IEDB catalogs all epitopes for humans and experimental animals for allergens, infectious diseases, autoantigens and transplants, and includes epitope prediction tools to accelerate immunology research around the world. Dr. Sette’s lab uses knowledge of epitopes to define the hallmarks of a beneficial immune response associated with effective vaccines, as opposed to immune responses that are ineffective or that cause harm. The laboratory’s infectious disease interests include SARS CoV2, dengue, Zika Chikungunya, herpesviruses, poxviruses, lassa fever, HIV and hepatitis viruses, and bacterial pathogens such as tuberculosis and bordetella pertussis. Our investigations outside infectious disease include allergic asthma and Parkinson’s disease.
Dr. Sette is a Doctor in Biological Sciences from the University of Rome and did postdoctoral work at the National Jewish Center for Immunology and Respiratory Medicine in Denver, Colorado. In 1988, Dr. Sette joined the newly founded company Cytel, in La Jolla, and was also appointed adjunct assistant professor at The Scripps Research Institute. He founded Epimmune in 1997, where he served both as Vice President of Research and Chief Scientific Officer until 2002, when he joined LJI as Head of the Division of Vaccine Discovery. He also heads the Center for Infectious Disease at LJI.

Questions addressed by the expert panelists include:
1. As viral immunologists you have all rapidly moved from your own area of expertise to characterising the immune response to SARS-CoV-2. What has most surprised and intrigued you in the study of the virus?
2. If SARS-CoV-2 variants totally evade vaccine-induced neutralising antibodies, can vaccine-induced memory T cells still contribute to host protection?
3. What insights do you have regarding the immune response in the context of re-infection of the same or different variants?
Our sincere thanks to the expert panel for their contribution:
Prof. Eui-Cheol Shin (KAIST, Korea)
Dr. Nina Le Bert (Duke-NUS Medical School, Singapore)
Prof. Tao Dong (University of Oxford, UK)
Dr. Yanchun Peng (University of Oxford, UK)

Dr Amy Flaxman is part of the Oxford Vaccine Group, Jenner Institute, University of Oxford. For her post-doctoral research she has worked on both pre-clinical studies developing vaccines for outbreak pathogens, such as Ebola, and clinical trials for outbreak pathogens, including MERS and Ebola. She is currently investigating immune responses to the Oxford ChAdOx1-nCoV vaccine for SARS-CoV-2 that was in Phase III clinical trials but is now approved. She leads the lab team carrying out antibody testing post-vaccination. Here, she is interested in the differences in antibody responses induced over time with different doses of vaccine, in different age groups and how this changes with administration of a booster vaccine.

Yanchun Peng is a senior Postdoctoral researcher working at the Human Immunology Unit, Weatherall Institute of Molecular Medicine, University of Oxford. After completing her undergraduate studies and then teaching in China, she moved the UK, first working as a research assistant, then completing her DPhil and remaining as a Postdoc in the research group led by Prof. Tao Dong since 2005. She has acquired extensive experience and skills in human T cell immunology with the major focus of her research being on evaluating anti-viral efficacy of virus-specific T cells and the interaction of host and virus in HIV infection, HIV/HCV co-infection and influenza infection. Her recent interest is to study the Viral OncoProtein(VOP) and tumor Specific Protein(TSP) specific T cell responses in virus associated cancer (ie HBV/HCC; EBV/NPC and HPV/CC) by combining cutting edge technologies (such as multi-colour Flow cytometry, Mass Cytometry (CyTOF), single cell RNASeq with SmartSeq 2 and 10X Chromium) with my expertise in isolating, expanding and characterising virus/tumor-specific T cells in vitro. Being equipped with these well-established platforms, skills and knowledge in human T cell immunology, she swiftly switched my work from cancer to COVID-19, as the pandemic started in 2020: exploring the role of SARS-CoV-2 -specific T cells in COVID-19 infection.

Since the early days of the development of the CRISPR Cas9 gene editing technology there have been sporadic speculations that immune responses to Cas9 (which is of microbial origin) could have consequences for clinical applications. Since late last year, three studies have presented experimental evidence for T- and B-cell responses to Cas9. These results have prompted considerable commentary, in both the scientific and popular press, marking Cas9-immunogenicity as a major obstacle to bringing CRISPR genome-editing to the clinic. The studies on Cas9 immunogenicity have brought attention to a critical element in the eventual clinical application of Cas9-mediated gene editing. In this presentation I will provide results of recent studies using a broad array of assays including ELISAs to detect anti-Cas9 antibodies, T-cell proliferation assays to identify immune-hot spots on Cas9 and MHC Associated Peptide Proteomics. The experimental data demonstrate the need for fit-for-purpose assays, statistical methods and reagents that will be necessary for obtaining data that is sufficiently reliable to make decisions for drug development and licensure. The broad outline of my presentation is as follow:
• Unique challenges while assessing the immunogenicity risk to Cas9
• The need of validated reproducible assays that are fit-for-purpose
• The need for well characterized reagents and for controlling contaminants and impurities
• Model systems for evaluating the immunogenicity of Cas9

Sequence variations in CMV UL40-derived peptides control NK activation through HLA-E presentation.

Oesophageal cancer (OC) causes over 0.5 million deaths worldwide each year. Current therapeutic regimens focus on chemo-radiotherapy prior to surgery, however, only 20-30% of patients respond to treatment. Therefore, new treatments are badly needed. Natural killer T (NKT) cells are innate T cells that recognize lipid antigens presented by CD1d molecules. We enumerated circulating NKT cells in OC patients and control subjects by flow cytometry using monoclonal antibodies and CD1d tetramers loaded with glycolipid antigens.
This allowed for the detection of invariant NKT cells, which possess invariant T cell receptor (TCR) α-chains reactive with α-galactosylceramide and mediate anti-tumour immunity in mice and humans, as well as novel NKT cells that recognise other glycolipids. We found that invariant NKT cells were depleted and functionally-impaired in patients with OC. In contrast, NKT cells reactive against sulfatide and tetramyristoyl-cardiolipin (TCL) were present in significantly higher numbers in OC patients compared to controls. Most of these cells expressed γδ TCRs, specifically the Vδ1 subset. TCL induced the production of transforming growth factor-β by expanded Vδ1 T cells in vitro in a CD1d dependant manner. Supernatants of Vδ1 T cells activated with sulfatide or TCL inhibited degranulation by invariant NKT cells. These data suggest that invariant NKT cells protect against OC but other CD1d-restricted NKT cells may promote tumorigenesis in these patients. These findings have important implications for cellular therapies involving invariant NKT cells, which are currently being tested in clinical trials for multiple cancer types.

Ankyrons™ are next-generation target binding reagents provided by ProImmune that overcome many of the current constraints of research antibodies. They are highly stable recombinant single-domain binders that can be directly labelled or detected via an epitope or biotin tag. Ankyrons can be engineered to combine them with further Ankyron or other domains much more easily than even recombinant Antibodies. Unlike Antibodies, Ankyrons are never made in animals and do not have isotypes or any species origin.

Defence Science and Technology Laboratories, UK
The Human Side of Counter-Bioterrorism: T Cell Proliferation Assays Validate Humanization of an anti-VEEV Antibody
Gene Olinger, USAMRIID
ProImmune’s Antigen Characterization Platform Accelerates Development of a Vaccine for Ebola and Marburg Viruses
Giovanna Rappocciolo, University of Pittsburgh
ProImmune’s REVEAL & ProVE® Rapid Epitope Discovery System Validates Novel Epitopes Important for CD8+ T Cell Control of Human Herpesvirus 8
Andre Oliveira, University College Dublin
ProImmune REVEAL and ProVE® Used to Identify First HTLV-2 MHC Class I Epitope
Pim van der Heiden, Leiden University Medical Center
Discovery of First VZV HLA-A2 Epitope, Using ProImmune REVEAL & ProVE® Technology
Katharina Steinitz, Baxter, Austria
What causes adverse reactions to Factor VIII treatment?
Julio Delgado, University of Utah
The Who How and Why of Seafood Allergy, using ProImmune’s Antigen Characterization Platform
Dolores Jaraquemada, Universitat Autònoma de Barcelona
Grave’s Disease Peptides are Characterized using ProImmune Class II REVEAL Binding Assay
| Philippe Blancou, INSERM Identification of New Epitopes in Type 1 DiabetesLena Israelsson, Karolinska Institute Defining Autoantibody Binding to Citrullinated Target Proteins in Rheumatoid Arthritis using Peptide Arrays |
 |
Kaare Engkilde, University of
Copenhagen
Mouse CD1d Tetramers Used to Correlate NKT Cell Activity with Protection from Type 1 Diabetes
Damien Bresson, LIAI
Research Into Type I Diabetes Therapy Makes use of PEPscreen® Technology
Terrance O’Hanlon, National
Institute of Environmental Health Sciences
Invited Editorial: Epitope Discovery for
Inflammatory Myopathy Treatment
More Autoimmunity
Publications Citing ProImmune

Immunovaccine Inc, Nova Scotia, Canada
Liposomes deliver an immune hit to cancer cells
Marcus Butler, Dana Farber Cancer Centre
Killer T Cells Trained for Long-lasting Anti-Cancer Effects
John Webb, British Columbia Cancer Agency
High-grade Ovarian Cancer: MHC Pentamers are Used to Investigate Immunotherapy Prospects
| Ingerid Abrahamsen, Oslo University Hospital, Oslo Allogeneic T Cells May Be Targeted to B Cell LeukemiaErlend Stronen, Radiumhospitalet Oslo Pentamers Identify Allo-restricted T Cells in Study to Advance Research Into Cancer Immunotherapy |
 |
Richard Harrop, Oxford Biomedica
Novel Epitope Detection in Phase II Trial for Colorectal Cancer
Maxim Pavlenko, Karolinska University Hospital
Epitope Discovery in Prostate Cancer: Characterizing an Immunodominant CTL Epitope of PSA in Mice
Vincenzo Cerullo, University of Helsinki
First Ever Tumor-specific Response Observed for Human Patients in Oncolytic Adenovirus Cancer Gene Therapy Study
James Wells, King’s College London
New Adjuvant Drives Unprecedented Cytotoxic T Cell Response Providing a Potent Vaccine Development Platform
Hiro Yagi, Hamamatsu University School of Medicine
Pro5® Pentamers Integral in Successful Research Into Novel Immunization Strategy
Norman Woller, Edkulla Ramakrishna, Medizinische Hochschule Hannover
Using thinkpeptides to Improve Virotherapy Against Tumor Targets
More Cancer Publications Citing ProImmune
| Claire Ventura, Sanofi Pasteur, France Finding a way to control chronic CMV infection Daniela Weiskopf, Austrian Academy of Sciences
Paul Moss, University of Birmingham
|
 |
Stephanie Gras, Queensland Institute of Medical Research
Judy Tellam, Queensland Institute of Medical Research
| Christopher Fox, University of Birmingham Researchers Use Pro5® MHC Pentamers to Identify an Exciting New Target for EBV Treatment Michael Uhlin, Karolinska Institute Pro5® Pentamers Central in Life-saving Procedure for PTLD Treatment Michael Uhlin, Karolinska Institute Invited Editorial: The Best Strategy for Adoptive T Cell Transfer |
 |
Suzanne Elliot, Queensland Institute of Medical Research
Pro5® MHC Class I Pentamers Used to Evaluate Vaccine Safety
John J. Miles, Cardiff University School of Medicine
Invited Editorial: Tracking the TCR Repertoire Evolution During Primary Viral Infection in Humans
More EBV publications citing ProImmune
Katherine High, The Children’s Hospital of Philadelphia
Pro5® MHC Pentamers used in conjunction with ProPresent® Antigen Presentation Assays to identify MHC class I epitopes in AAV vectors that trigger cytotoxic T cell responses when presented by hepatocytes
Adam Gehring, Singapore Institute for Clinical Science
Phenotypic Analysis of HBV-specific T Cells Using Pro5® Pentamers
Victoria Kasprowicz, University of KwaZulu Natal
A Story of Viral Escape Through Sequence Variation in Chronic Hepatitis C Virus Infection
Lars Frelin, Karolinska Institute
Developing a Therapeutic Hepatitis B Vaccine with Pro5® Pentamers
Eleanor Barnes, University of Oxford
Success for Hepatitis C Vaccine Trials at Oxford University
Tanja Lange, University of Lübeck
Sleep Your Way to Better Immunity – Sleep After Vaccination Boosts Immunological Memory
Samantha Westrop, Chelsea & Westminster Hospital
ProImmune REVEAL & ProVE® Rapid Epitope Discovery System used to Identify Novel T Cell Epitopes in HIV-1
Jorge Almeida, INSERM
T Cell Vaccines for HIV-1 Move a Step Closer – Researchers Use HIV-specific Pro5® MHC Class I Pentamers Combined with Intracellular Cytokine Staining
Aude Chapuis, Fred Hutchinson Cancer Research Center, USA
Sending in Trained Killers to Attack Residual HIV Infection
University of Manitoba Researchers Use PEPscreen® Library for Epitope Mapping
Tarmo Mölder, University of Tartu
HIV Vaccine Study in Pigs Uses PEPscreen® Peptides in ELISpot Assays
More HIV Publications Citing ProImmune
Infectious DiseasesPaul Arnaboldi, New York Medical Center Sanja Selak, Intercell AG Maziar Divangahi, Harvard Medical School, USA Ana Ferreira, Instituto Gulbekian de Ciência, Portugal |
 |
Virginie Roupie, Scientific Institute of Public Health, Brussels
PEPscreen® Used in Vaccine Development for Mycobacterium Tuberculosis
Cyrille Bisseye, Medical Research Council UK and The Gambia
Designing a Malaria Vaccine

J Guo, University of Texas
Deletion of FoxN1 in the thymic medullary epithelium reduces peripheral T cell responses to infection and mimics changes of aging
Roberto Mallone, Hôpital Saint Vincent de Paul
How Does Blood Processing Affect the Results of Cell Mediated Immunity Assays? ProImmune’s Flu Pro5® MHC Class I Pentamer is Used as an Exemplar in an Exploratory Study

| Patricia Barral, Cancer Research UK ProImmune CD1d Tetramers Provide Novel Insights Into NKT Cell Activation Stefan Porubsky, DKFZ The Significance of Isoglobotrihexosylceramide (iGb3) as the Natural CD1d Ligand in Mice Maya Fedeli, H San Raffaele Scientific Institute NKT Cell Differentiation Depends on Dicer Processing of MicroRNA |
|
Thierry Mallevaey, INSERM
NKT Cells are Important in the Acquired Immune Response During Helminthiasis
Omar Duramad, Regimmune
ProImmune’s Human CD1d Tetramer Detects NKT Cells from Non-human Primates

Narasimha Rao (Dr Reddy’s Laboratories, India)
“At Dr. Reddy’s Laboratories, we have had the privilege of collaborating closely with ProImmune for immunogenicity risk assessment services. Their team demonstrates best-in-class expertise in evaluating complex peptide products, providing critical insights that have been invaluable to our development process. What truly sets ProImmune apart is their exceptional professionalism and commitment to supporting regulatory timelines. Their rigorous approach and high-quality services directly contributed to our ability to file peptide products with regulators efficiently. Working with ProImmune has been a consistently positive experience – their scientific excellence, combined with their understanding of regulatory requirements, makes them an ideal partner for any organization developing complex peptide therapeutics.“
Annelie Hellvard (The Gade Institute, University of Bergen, Norway)
“We have had nothing but good experiences using peptides synthesized by ProImmune/thinkpeptides. Since we started working with them in 2009 there have been no issues whatsoever, and the peptides were a big part of a recent publication in the Journal of Immunology [PubMedID:20488785]. I am always surprised by how fast quotes are delivered and questions are answered – the customer service is excellent. We feel like they have a very good product for decent price.”
Cecilia Rietz, Ph.D. (Lovelace Respiratory Research Institute, New Mexico, USA)
“We are using peptide pools to develop flow cytometric methods to assess T cell response in the quest for correlate/s of protection in animal models for development of vaccines against filovirus. The interaction with ProImmune has been integral to the good results we have had in the assay development thus far. From the design of the peptide pools, through the quote- and purchasing process and finally the reconstitution and use of the peptides in the assay, ProImmune is always there to answer questions with prompt replies, honesty and knowledge. In case of any unexpected events during the manufacturing process, we were always promptly notified and given options for how to proceed. ProImmune also showed a lot of flexibility during the design- and quote stages which we especially appreciate as a contract research organization. As for the quality and consistency of the peptide pools in the assay, we are completely satisfied. I would highly recommend working with ProImmune to my colleagues and I would definitely work with them again on similar assay development projects.”
Dr. Vesna Blazevic (Vaccine Research Centre, University of Tampere, Finland)
“ProImmune’s PEPscreen® library has played a central role in my research into DNA based and HIV vaccines in both IFN gamma and IL-2 ELISpot assays. Working in preclinical and clinical settings, I have found the library to be an easy and reliable way of screening for T cell epitopes. The peptides were easy to dissolve, of a very good quality and showed no non-specific background in the ELISpot assay. My decision to purchase products from ProImmune was based on the fact that they are a good price, of a superior quality and have fast delivery. I look forward to using ProImmune’s products in my future research.”
Dr. Yared Haliemichael (University of Texas MD Anderson Cancer Center, USA)
“I use the B16 melanoma/pmel-1 T cell model to study the requirements for successful vaccination using peptide immunogens. The induction of strong CD8 T cell response requires a sustained presentation of antigen in a stimulatory context to develop into effector cells that can kill tumors through the recognition of tumor antigens bound to MHC Class I molecules. It is important to have a high standard of reagents for my mouse vaccination protocols, and I am very satisfied with the quality of peptides and the responsive customer service that thinkpeptides provides.” (thinkpeptides is ProImmune’s brand for custom peptide synthesis services)
Dr. Karen Fitzmaurice (Nuffield Department of Clinical Medicine, University of Oxford, UK)
“Our study aimed to characterize the CD8+ T cell responses associated with certain class I alleles linked to favourable outcomes in HCV infection. We chose a PEPscreen®: Custom Peptide Library of overlapping peptides spanning the HCV genome to use in ELISPOT assays. As well as the fact that the peptide library was of good quality and easy to reconstitute for use in the assays, I also found the price to be competitive and the service I received was excellent.”
Dr. Arun Rishi (John D. Dingell VA Medical Center, Detroit, USA)
“I have been consistently impressed by the customer service I received from thinkpeptides, as well as the quality of their products. They pay great attention to detail regarding my shipping requirements, are very easy to talk to, and are always very professional. I will certainly continue to order my peptides from thinkpeptides.”
Dr. Johannes Wiegand (University of Leipzig, Germany)
“My research is investigating the T cell immunology of viral hepatitis. Using PEPscreen® overlapping peptides from ProImmune in IFN gamma ELISpot and 3H-proliferation assays, enabled us to map the entire Hepatitis B surface antigen and HBV-core antigen region for T cell analysis. ProImmune was personally recommended to me by Dr. Heiner Wedemeyer and true to his recommendation, I found their peptides to be of a very good quality and easy to use. The administrative team were very helpful with the process of billing and shipping and I had good communication with my account manager who made useful comments and was highly motivated to solve any issues.”
Dr. Norman Woller (Hannover Medical School, Hannover, Germany)
“Our laboratory has used ProImmune peptides (thinkpeptides) for years in studying viruses and tumour-specific immune responses. Furthermore, we are using custom synthesised Pro5® Pentamers for detection of tumour-specific CD8 T cells and are highly satisfied with the results. We appreciate the high quality of each individual batch and we consider ProImmune as the provider of choice for tools to reliably investigate cellular immunity.”Read his full testimonial, including information about his career and publications here.“
Dr. Peng Li (Eisai, Japan)
“From our first contact with ProImmune in October 2022, they were very responsive and efficient in their communication where they quickly addressed all our enquiries via email and online meetings, enabling us to make a rapid decision to proceed with our DC-T cell assay project within just one month. What truly stood out was their exceptional turnaround time – the entire project, involving testing of 11 proteins against 42 HLA-typed healthy donors, was completed in approximately six weeks following paperwork finalization and shipment of materials to their UK facility. Beyond the speedy execution, ProImmune demonstrated outstanding scientific support. They scheduled a comprehensive debrief call to help interpret the data and answer our questions, which proved invaluable in helping us reach our project milestones.“
Dr. Caroline Barelle (University of Aberdeen, Scotland)
“We are developing shark single domain therapeutics so identifying and addressing any immunogenicity issues is a critical part of our pre-clinical candidate selection process. Our initial discussions with ProImmune were incredibly informative and helped us define the experimental approach we should adopt. They provided support and information throughout to keep us updated and also succeeded in delivering the data package within our desired and indeed aggressive time frame. In addition, they provided follow up support to take us through the final report to ensure we had a clear understanding of the results obtained. Overall we are very happy with the service and would work with ProImmune again.“
Prof. Eui-Cheol Shin (Korea Advanced Institute of Science & Technology, South Korea)
“ProImmune is one of the most outstanding providers of MHC Class I Multimers. Beyond their catalog of high-quality MHC Class I Pentamers, they have consistently delivered exceptional custom synthesis services tailored to my epitope requirements. Their team provided clear communication and expert guidance to design optimal MHC Class I Pentamers, which were critical to the success of my research. During the early stages of the COVID-19 pandemic, ProImmune demonstrated remarkable agility by rapidly synthesizing a broad range of SARS-CoV-2-specific MHC Class I Pentamers. Thanks to their fast turnaround time, I was able to obtain these essential reagents quickly. I ordered nearly 10 very diverse Pentamers and it worked very successfully—enabling my team to publish one of the first papers characterizing SARS-CoV-2-specific CD8+ T cells in infected patients. This work marked the beginning of my journey in SARS-CoV-2 immunology, and ProImmune played a pivotal role in that achievement.
One of the greatest advantages of working with ProImmune is their collaborative approach. Their Asia-Pacific representative keeps me updated on the latest reagents and protocols, ensuring that I’m not just purchasing a product but also receiving valuable scientific support. Before placing an order, I can consult with their specialists to discuss the best experimental protocols and reagents for my project.“
Dr. John Webb (British Columbia Cancer Agency, Canada)
“Over the past six years I have been using ProImmune’s Pro5® Pentamers for the identification and characterization of tumour epitope-specific T cells. Pentamers play an important role in my work to develop new vaccine strategies to induce T cell-mediated immune responses directed against human malignancies. I am always impressed by the knowledge and friendliness of the customer service team, and find the website very user-friendly and well-organized.”
Prof. Dirk Schlueter (Otto-von-Guericke Universitaet, Magdeburg, Germany)
“I have used ProImmune’s Pro5® MHC Pentamers in my immunology research into infectious diseases in murine models. Using the Pentamers in flow cytometry, I have found them to be of good quality and easy to use. Over the past 3 years I have had a positive experience with the company and have found them to be competitively priced and swift to manufacture and deliver their products“.
Dr. Georg Lauer (Harvard Medical School, Boston MA, USA)
“The use of peptide-coupled MHC multimers allows us to identify very rare specific cell populations with high sensitivity and specificity by flow cytometry. This was only possible for several advantages of the ProImmune Pro5® MHC Pentamer technology. Because the HCV T cell response repertoire is highly diverse, we need to have more than 20 different HCV multimers in stock in order to study a majority of our subjects. The wide range of HLA class I alleles on offer but also the opportunity to freeze the unlabelled pentamers has greatly facilitated this work. We also use many novel antibodies that are only available in a limited range of fluorochromes and here the unlabeled pentamers with the option of using different fluorotags has allowed us much more flexibility in designing our 10 color flow panels.Overall class I peptide multimers have greatly enhanced our ability to analyze human CD8 T cell responses directly ex-vivo with unprecedented specificity and sensitivity, allowing us for the first time to perform human studies that are a serious complement to studies of viral immunity in murine models.”
Dr. Daniel Volker (Institute of Immunology, Heidelberg University, Germany)
“We use ProImmune Pentamers specific for HIV epitopes to enable us to monitor the specificity and number of CD8+T cells in our cohort of patients over time. The range of epitope specificities and the high quality of reagents we obtain from ProImmune are key to the success of our work. At the moment we are really excited, as we are on the verge of publishing our findings regarding CD8+ T cell numbers in HIV+ individuals, and how these correlate with a prognosis for AIDS development. This work has been made possible by ProImmune.”
Dr. Cath Bollard (Baylor College of Medicine, Houston TX, USA)
“Pro5® MHC Pentamers have played an important part in our study on adoptive immunotherapy for cancer and viral infections, post-transplant. We use Pentamers to detect and quantify the populations of antigen-specific T cells in CTL lines made for clinical use. We also use them to detect antigen-specific CTL in the peripheral blood of transplant patients, pre and post-CTL-infusion. The Pentamers were chosen for their stability and for the increased intensity and specificity of the positive population compared to MHC tetramers. We have found Pentamers to be consistent and reliable, and ProImmune’s customer service is excellent“.
Prof. Thomas Brocker (Institute for Immunology, University of Munich, Germany)
“ProImmune Pro5® MHC Pentamers have been influential in our research to analyze CD8 T cell responses in vivo. Colleagues in the lab recommended the Pentamers to us. For the past two years we have used them for flow cytometry applications to achieve reliable staining across different batches of the same product. I can also recommend ProImmune for their easy ordering process and excellent customer service.“

Dr. Navapon Techakriengkrai (Chulalongkorn University, Thailand)
“I outsourced my tissue typing to ProImmune because they provide me with the fastest service at the best price. Since the main theme of my research is on T cell immunology, HLA-type is inevitably needed. The customer service from ProImmune is of the best quality. Their typing service allowed me to unambiguously resolve the HLA type of many of my samples.”
Dr. Wivine Burny (GSK Bio, Rixensart, Belgium)
“After being in contact with the friendly sales team at ProImmune, I decided to outsource my tissue typing and take advantage of their type HLA service. For us, a major advantage of the service was the competitive pricing, and we could ask for a small number of samples to be typed at once. The typing was performed rapidly and results were delivered as promised: punctually and in a clear report. The team at ProImmune were prompt in responding to the additional questions I had about my report.”
Dr. Jennifer Kirchherr (Duke Human Vaccine Institute, North Carolina, USA)
“We have a programme of clinical studies and it is important for our researchers to be able to correlate the immune responses they observe with HLA type. We looked at a range of companies offering tissue typing, and also at using an on-campus typing facility, and we found that ProImmune were able to offer us the best pricing and turnaround time. We now send them regular batches of samples to type. They have been wonderfully easy to work with, if we ask anything they get right back to us and overall it has been a really good experience.“

Dr. ShuTing Han (National Cancer Centre Singapore, Singapore)
“My experience working with ProImmune has been exceptionally positive. What truly differentiates them is their expert guidance in designing peptides for custom Class II tetramer synthesis – a level of scientific partnership I haven’t found with other vendors, including core facilities like those at NIH. While other providers simply fulfil requests with numerous caveats and long wait times, ProImmune’s team actively collaborates with researchers throughout the entire process. Their ability to deliver custom Class II tetramers in a timely manner, combined with their hands-on scientific support.”
Dr. Mitchell Zheng (University of Melbourne, Australia)
“At the forefront of vaccine development, our research focuses on decoding the mechanisms of murine and human CD4+ T follicular helper (Tfh) cells. These investigations demand the highest quality reagents – a need consistently met by ProImmune. For many years, we’ve relied on ProImmune’s comprehensive MHC class II catalog and custom monomers, including pre-conjugated variants, with our lab ordering a diverse range of specialized reagents. This sustained partnership, expertly facilitated by ProImmune Asia-Pacific representative, stands as clear evidence of our trust in ProImmune’s reliability and technical excellence.
A recent challenge in mapping novel CD4+ T cell epitopes required custom tetramers capable of tracking antigen-specific cells in their native state. While other vendors fell short, ProImmune delivered where others couldn’t. Their successful development of this critical reagent has opened new avenues in our research program. Working with the ProImmune Asia-Pacific representative has been exceptional. Their specialist combines deep immunological expertise with remarkable responsiveness, enabling rapid procurement of precisely tailored reagents. What truly distinguishes ProImmune is their scientific partnership approach. When theoretical models suggested a particular monomer synthesis would fail, they carefully evaluated our empirical staining data and proceeded with production. This data-driven, collaborative mindset has repeatedly accelerated our discoveries, making ProImmune not just a supplier, but a true research enabler.“
Dr. Michael Mathis (Louisiana State University Health Sciences Center, USA)
“My work focuses on the development of adenovirus-based vectors for immunotherapy of breast cancer, using mouse models of the disease. ProImmune’s REVEAL® Rapid Epitope Discovery System has provided a cost-effective method for me to screen my proteins of interest for potentially immunogenic epitopes. I have been particularly impressed with the service that ProImmune offers, so much so that I’ve been telling my colleagues that they too should outsource more of their routine experiments to experts in order to free up their time. It really saves time and money when one take into account the effort needed to set up these assays in house and then wait for results. I will definitely be using the system again to obtain further data on the hits from the first round of screening.”
Dr. James Wells (Beth Israel Deaconess Medical Center, Boston, USA)
“I have dealt with ProImmune in the past and they have consistently proven to provide a high level of service. ProImmune’s REVEAL® service provided me with the option to test the in vitro binding properties of a selection of my most promising peptides quickly and efficiently, resulting in data I know I can trust.I have always found ProImmune to be very approachable and reliable, and their willingness to make contact with me directly over the telephone (following my initial e-mail request for further information) to discuss my requirements greatly helped me to fully understand the services offered and reassured me that my analysis would be conducted in a professional manner. The quotation process was simple, and my results were presented in several different formats to allow me to easily determine which peptides stood out for further investigation. Based on my results I have since gone on to order my custom peptide synthesis through ThinkPeptides, a branch of ProImmune.”
Dr. Ruben Varela-Calvino (University of Santiago de Compostela, Spain)
“Dr. Varela-Calvino has been researching a protein which acts as a target in autoimmune diabetes, and recently benefited from the ProImmune REVEAL® HLA-peptide binding assays. He and his team wanted to analyze the binding of some candidate peptides from his protein to HLA-DRB1*04:01, one of the major HLA types associated with autoimmune conditions. Dr Varela-Calvino has extensive experience performing HLA-peptide binding assays, and, recalling the length of the process, it wasn’t an experience he was keen to repeat: “Instead, I performed an internet search, and found ProImmune. I didn’t want to ask members of my lab to spend years optimizing an assay which ProImmune runs routinely – time in the lab is precious, particularly for Ph.D students”.ProImmune provided HLA-peptide binding assay results in a matter of weeks. His team were quickly able to move on to validating the binding assay data from ProImmune, and were delighted to find that the data they obtained from ELISpot experiments on patient samples correlated almost exactly with their results. As he moves towards collating his data for publication, Dr Varela-Calvino has nothing but praise for ProImmune. He told us “I’ve found the service from your team highly professional, the turnaround time for assays very impressive, and I would certainly work with ProImmune again in future”.
Dr. Birgit Schultes (Unither Pharmaceuticals, Massachusetts, USA)
“The report provided with the service and reagents was well organized and easy to understand. We are very happy with the data, the technical support and the on-time delivery of this technology. Several peptides and Pentamers were identified that will be useful for immune monitoring. Those Pentamers are currently confirmed with patient samples, pre- and post OvaRex® treatment”
Minor Histocompatibility Antigen (miHAg)
ELISPOT Assay
Mycobacterium Tuberculosis
B cell Epitope Discovery Services
Respiratory Syncytial Virus (RSV)

Jambari, N. et al (2021) Effect of O-linked glycosylation on the antigenicity, cellular uptake and trafficking in dendritic cells of recombinant Ber e 1. PLoS One 16: 10.1371/journal.pone.0249876 [PubMedID: 33914740]
Steven, J. et al (2017) In Vitro Maturation of a Humanized Shark VNAR Domain to Improve Its Biophysical Properties to Facilitate Clinical Development. Frontiers in Immunology. [PubMedID: 29109729]
Trypanosoma Cruzi
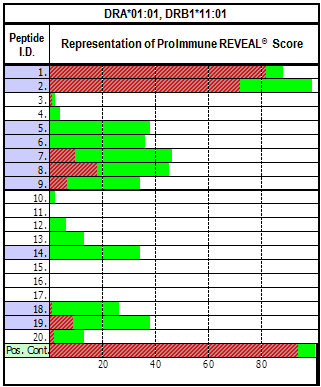
Vaccinia Virus
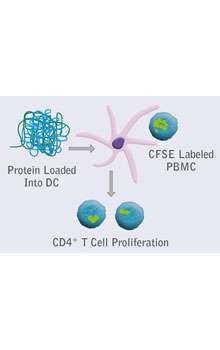
Other

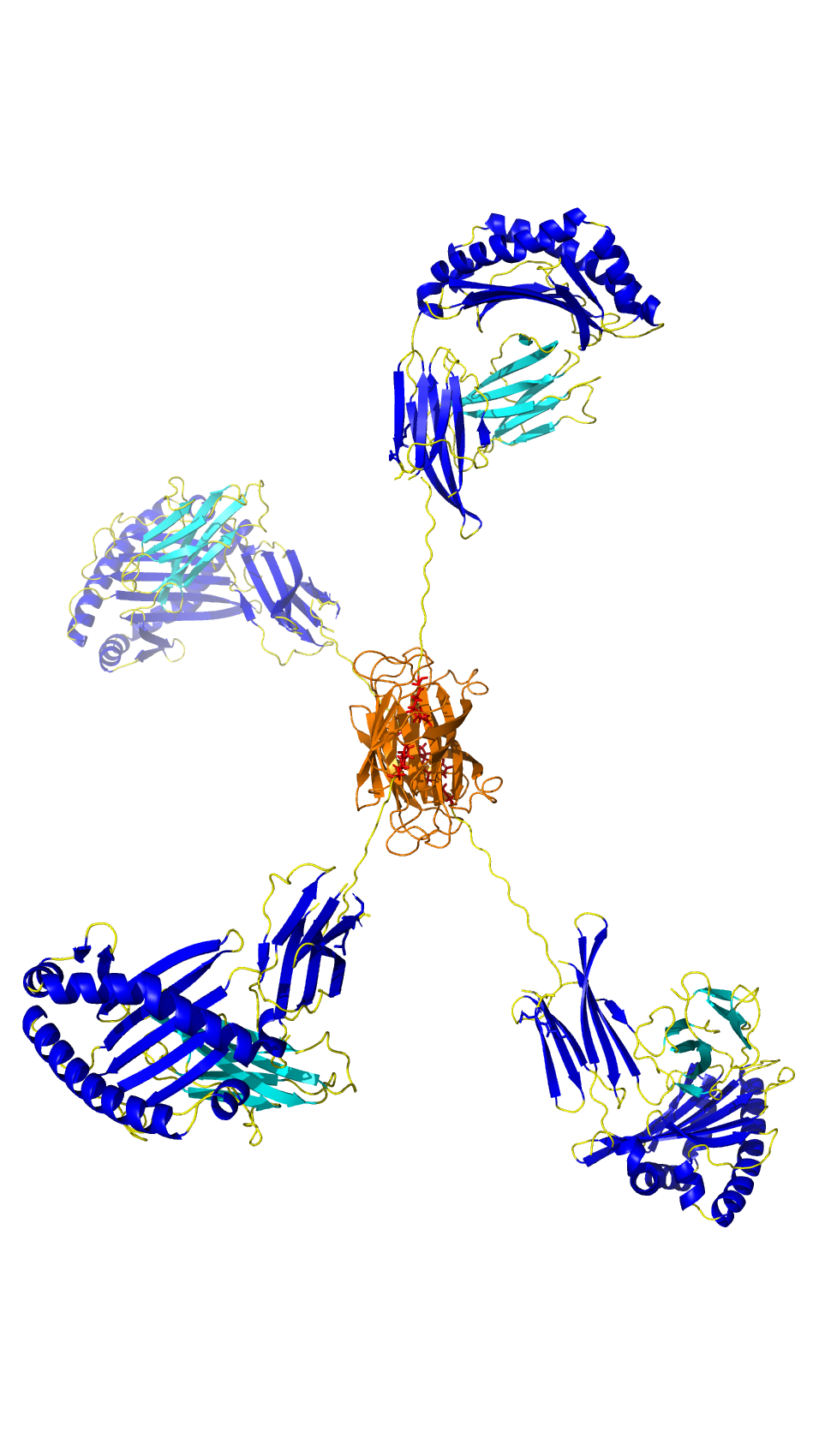

Kramer C. et al. (2020). “Generation and reactivity analysis of human recombinant monoclonal antibodies directed against epitopes on HLA-DR” AJT DOI: 10.1111/ajt.15950
Hartnell F. et al. (2020). Characterising HCV specific CD4+ T-cells following viral-vectored vaccination, directly acting anti-virals and spontaneous viral cure. https://doi.org/10.1002/hep.31160
Cianciotti B. et al. (2019). CD4+ memory stem T cells recognizing citrullinated epitopes are expanded in patients with rheumatoid arthritis and sensitive to tumor necrosis factor blockade. https://doi.org/10.1002/art.41157
Kramer C. et al. (2020). Generation and reactivity analysis of human recombinant monoclonal antibodies directed against epitopes on HLA-DR. https://doi.org/10.1111/ajt.15950
Gliwinski M. et al. (2020). Proinsulin-specific T regulatory cell may control immune responses in type 1 diabetes: implications for adoptive therapy. 10.1136/bmjdrc-2019-000873
ProImmune Peptides

ProMix™ Peptide Pools
Malaria
ProPresent® Antigen Presentation Assay

