Your basket is currently empty!
think peptides
the source for all peptides for your research
Custom Peptides
Peptides play a key role in many biochemical and immunological assays, including enzyme-substrate studies, blocking and competition assays, epitope mapping, T cell stimulation and many more. We bring you a custom peptide synthesis service that recognizes the importance of high quality reagents for these applications, delivered to you with a fast turnaround time and at excellent value.
Custom Peptide Synthesis Product Range
Our standard peptide synthesis service includes the following purities and quantities of peptide. Contact us if you require lower grade purity or larger amounts, we can also supply in gram quantities.
|
Purity |
>70%, >75%, >80%, >85%, >90%, >95%, >98% |
|
Quantity |
1-4 mg, 5-9 mg, 10-14 mg, 15-19 mg, 20-24 mg, 25-29 mg, 50-59 mg |
|
Typical dispatch time |
>70% and >75% purity, approximately 3 weeks |
If your requirements fall outside the range of scales, purities listed above, contact us for a special quotation tailored to your needs.
Alongside standard custom peptide synthesis and a wide range of modification options, we offer high-throughput synthesis of custom peptide libraries. Peptide libraries can be used to investigate immune responses in detail or can provide an efficient method of screening the activity of a large number of peptides in an application.
Expert advice working with peptides
To get the most from peptides in your experiments, you need to consider all relevant factors when designing and using peptides. Our peptide eBook provides expert advice on the storage and handling of custom peptides, to enable you to get optimal results. It covers the implications of the presence of certain amino acids in the peptide sequence and suggested design consideration to help achieve your desired outcome.
|
|
Download the thinkpeptides eBook
Key publication on use of thinkpeptides in clinical trial evaluation of Oxford COVID-19 vaccine:
Folegatti P., Ewer K., et al., on behalf of The Oxford Covid Vaccine Trial Group
The Lancet (2020) Volume 396, ISSUE 10249, P467-478 https://doi.org/10.1016/S0140-6736(20)31604-4.
“Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial”
Adapted abbreviated summary:
The safety, reactogenicity, and immunogenicity of a viral vectored coronavirus vaccine (ChAdOx1 nCoV-19) that expresses the spike protein of SARS-CoV-2 was assessed.
There were no serious adverse events related to ChAdOx1 nCoV-19. In the ChAdOx1 nCoV-19 group, spike-specific T-cell responses peaked on day 14 (median 856 spot-forming cells per million PBMC, determined by ELISpot). Anti-spike IgG responses rose by day 28 and were boosted following a second dose. Neutralising antibody responses against SARS-CoV-2 were detected in all participants after a booster dose.
ChAdOx1 nCoV-19 showed an acceptable safety profile, and homologous boosting increased antibody responses. These results, together with the induction of both humoral and cellular immune responses, support large-scale evaluation of this candidate vaccine in an ongoing phase 3 programme.
Peptides used in this study:
258 15-mer peptides supplied as pools were provided by ProImmune (thinkpeptides) and used in IFNg ELISpot assays. Full details of the peptide list and sequences is published starting on page 23 of the supplemental information.
Key publication:
| Epitope map of Israeli Acute Paralysis Virus could help save declining honey bees |  |
Ren J. et al. (2014), “Assembly of Recombinant Israeli Acute Paralysis Virus Capsids.” Plos One doi:10.1371/journal.pone.0105943.g003
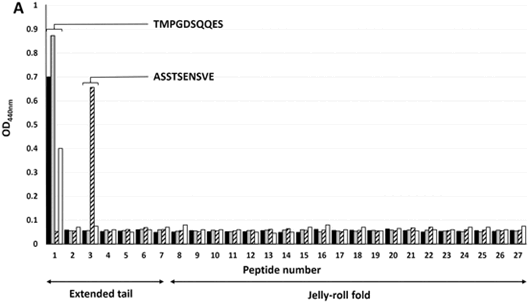
Figure 1. Linear antibody epitope map of 4 monoclonal antibodies to IAPV VP2 on an overlapping peptide library.
Summary: Israeli Acute Paralysis Virus (IAPV) has been implicated in the worldwide decline of honey bees. Ren et al studied the molecular protein biochemistry of how IAPV expresses and assembles into virus particles in insect cells. Monoclonal antibodies developed by Ren et al. specific to IAPV capsid proteins and an understanding of the highly specific epitopes they recognize as determined by Ren et al. could contribute to diagnostic tests to detect this bee pathogen. Such tests could play an important role in developing a better understanding of the epidemiology of IAPV in the honey bee population. Peptides for epitope mapping for this study were custom synthesized by thinkpeptides.
Praise for thinkpeptides from
Annelie Hellvard
The Gade Institute, University of Bergen, Norway
“We have had nothing but good experiences using peptides synthesized by thinkpeptides. Since we started working with them in 2009 there have been no issues whatsoever, and the peptides were a big part of a recent publication in the Journal of Immunology [PubMedID:20488785]. I am always surprised by how fast quotes are delivered and questions are answered – the customer service is excellent. We feel like they have a very good product for decent price.”
Modified Peptides
Our extensive range of custom peptide modifications includes, but is not limited to those in the table below. Contact us if you need a modification that is not on the list.
|
N-terminal |
Acetylation |
|
C-terminal |
Amidation |
|
Internal |
Biotin |
|
Non-standard residues |
Ornithine Norleucine Norvaline Homoserine Homocysteine Penicillamine Cyclization D-amino acidsN-methyl amino acids |
|
Other modifications |
Cyclization Glycosylation |
Design and Application Support from ProImmune’s Experts
Our technical and applications support team can offer advice on library design, peptide solubilization, experimental set-up, and analysis. Relating to design, we can advise on the best overlap or offset of peptides to be used in your target application, and then using your full-length protein sequence, we can generate the list of peptides for your library.
Custom Peptide Libraries
Synthetic custom peptides offer a rational, scaleable and ever more affordable approach for exploring protein-protein interactions or even more complex phenomena such as immune responses directed against specific epitopes. As the use of such peptides increases it becomes ever more important to consider all relevant factors when designing large peptide libraries for synthesis. Our applications scientists have the experience to help you get the peptide design right for your project. Our approach is to offer you flexible, scaleable synthesis wherever you are.
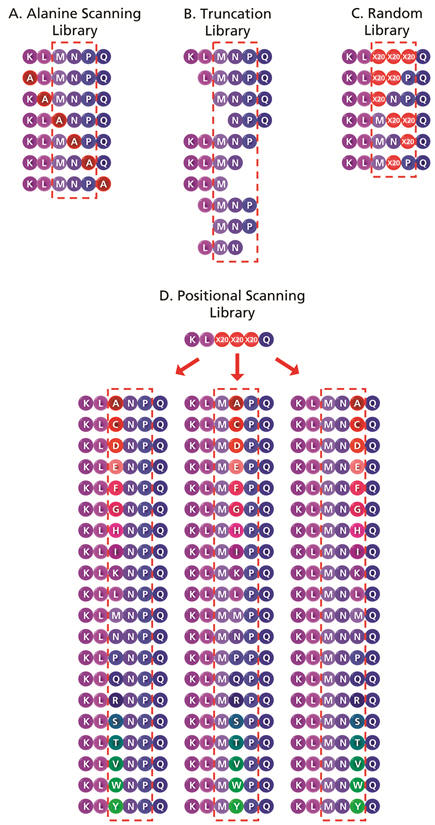
Figure 2. Schematic representation of the different strategies in constructing peptide libraries for sequence optimization. The presumed essential positions are enclosed in the dotted box. A. Alanine Scanning Library. Alanine is systematically substituted into each amino acid position in the identified epitope. This strategy identifies the amino acids in the native sequence that are essential for activity. Substitution of an essential amino acid results in a reduction in peptide activity, and the degree of reduction in activity is usually taken as a relative measure of the importance of the amino acid being substituted. B. Truncation Library. This strategy determines the minimum length required for optimum peptide activity by generating a set of peptides with systematic truncation of the flanking residues. If the essential amino acids are known, the direction of truncation can be selected around them, as opposed to systematic truncation from both ends of the peptide sequence. C. Random Library. Selected residues in the peptide sequence (wobbles) are simultaneously substituted with a mixture of all 20 amino acids, or a mixture of specific amino acids. In practise, this strategy is usually used for preliminary identification of a group of active sequences that can then be re-synthesized to validate the initial results. D. Positional Scanning Library. A selected position or positions in a peptide sequence are each systematically replaced with different amino acids in order to determine the preferred amino acid residues at these positions, measured by corresponding increases in activity.
Further key publications using thinkpeptides
Key publication:
thinkpeptides used in study to show antigen loading of tolerogenic DCs (tolDCs) reduces their capacity to induce tolerance in diabetes model
Funda D. et al., “Antigen Loading (e.g., Glutamic Acid Decarboxylase 65) of Tolerogenic DCs (tolDCs) Reduces Their Capacity to Prevent Diabetes in the Non-Obese Diabetes (NOD)-Severe Combined Immunodeficiency Model of Adoptive Cotransfer of Diabetes As Well As in NOD Mice” Frontiers in Immunology (2018), Vol 9, Art 290
https://doi.org/10.3389/fimmu.2018.00290
Abbreviated summary:
Antigen-unloaded tolDCs consistently decreased diabetes transfer in the NOD-SCID model of adoptive cotransfer of diabetes. On the other hand, tolDCs loaded with GAD65, with GAD65 immunodominant peptide no. 35 (synthesized by thinkpeptides), but also with a control protein OVA, all decreased their diabetes-preventive properties.
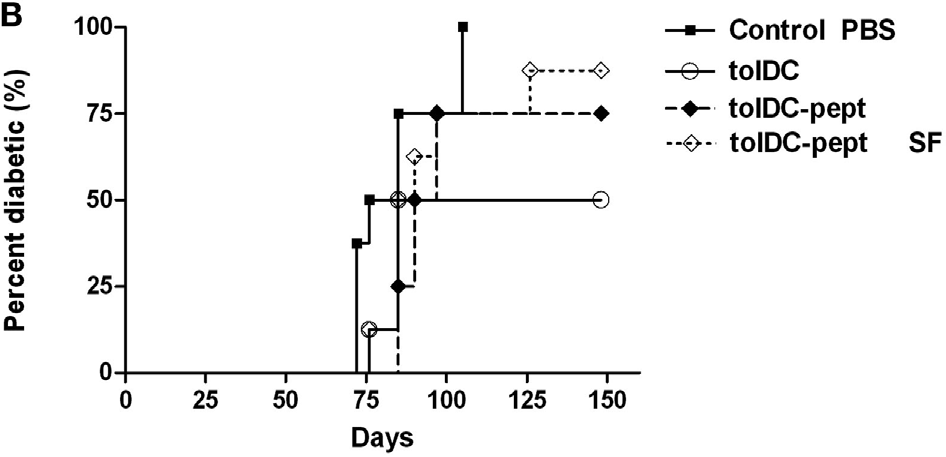
Figure. Unloaded tolDCs were compared to tolDCs loaded with 1 µg/mL of the GAD65-immunodominant peptide no. 35 (tolDC-pept) and prepared in both serum-supplemented and SF media. Diabetogenic splenocytes (5 × 106 per mouse) from 12-week-old prediabetic NOD females (n = 10) and above listed groups of tolDCs (3 × 106) were mixed and applied i.p. to 7-week-old NOD-SCID female recipients (n = 8). Diabetogenic splenocytes in PBS were used as the control group in both experiments. Data are presented as cumulative diabetes incidence in NOD-SCID recipients and p- values were compensated for multiple comparisons.
Key publication:
thinkpeptides used in study to show antigen loading of tolerogenic DCs (tolDCs) reduces their capacity to induce tolerance in diabetes model
Lim E.L. et al., “Phosphoinositide 3-kinase δ inhibition promotes antitumor responses but antagonizes checkpoint inhibitors”
JCI Insight (2018), Jun 7: 3(11)
doi: 10.1172/jci.insight.120626
Abbreviated summary:
The study demonstrates that PI3Kδ inactivation attenuates Treg suppressive function, which can unleash a potent antitumor immune response, conditional upon the dependence of the specific tumor type on Treg immunosuppression. However, the results highlight that the mechanism by which PI3Kδ inhibition leads to enhanced antitumor responses is distinct, and not necessarily complementary to that elicited using antibodies against CTLA-4 or PD-L1.
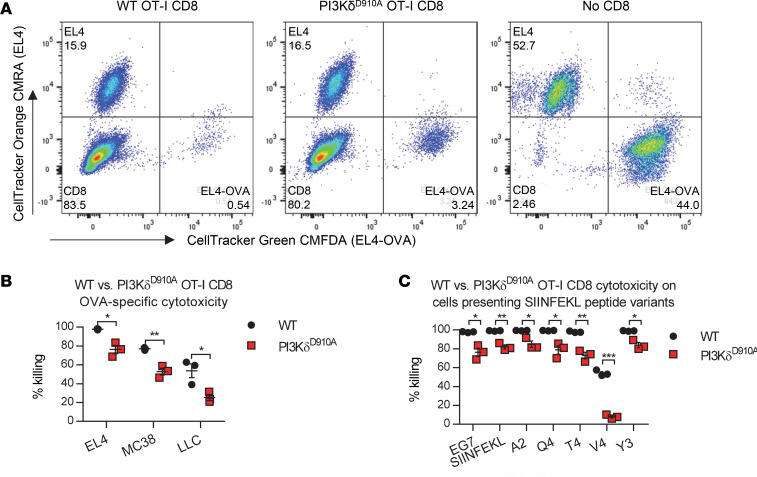
Figure. PI3Kδ(D910A) CD8+ T cells show functional defects. In vitro assays were conducted with 3 biological replicates. (A) CD8+ T cell cytotoxic capacity was measured in an in vitro killing assay, using tumor cells expressing ovalbumin (OVA) protein (stained with CMFDA CellTracker Green) as targets for in vitro–stimulated OT-I CD8+ T cells, and parental tumor cells (stained with CMRA CellTracker Orange) as controls. Representative FACS plots shows EL4-OVA and EL4 cells incubated with, from left to right, WT, PI3KδD910A, or no CD8+ T cells. (B) Cytotoxic efficiency measured in WT versus PI3KδD910A OT-I CD8+ cells using EL4-OVA, MC38-OVA, and LLC-OVA cells. Peptides for in vitro stimulation were synthesized by thinkpeptides.
Key publication:
thinkpeptides used in study to investigate lipid coated liquid crystal droplets for the on-chip detection of antimicrobial peptides
Bao P. et al., “Lipid coated liquid crystal droplets for the on-chip detection of antimicrobial peptides” Lab Chip (2019), Vol 19, 1082-1089,
DOI 10.1039/C8LC01291A
Abbreviated summary: A novel liquid crystal (LC) biosensor is used to demonstrate the detection of Smp43, a model antimicrobial peptide (AMP) from the venom of North African scorpion. Mono-disperse lipid-coated LC droplets were generated using microfluidic devices as targets for AMPs. The droplets were trapped and treated with Smp43 at gradient concentrations. The disruption of the lipid monolayer by the Smp43 was detected at concentrations well within its biologically active range, indicated by a dramatic change in the appearance of the droplets. This suggests the system has feasibility as a drug-discovery screening tool and the potential for development as a reliable, low-cost and disposable point of care diagnostic tool.
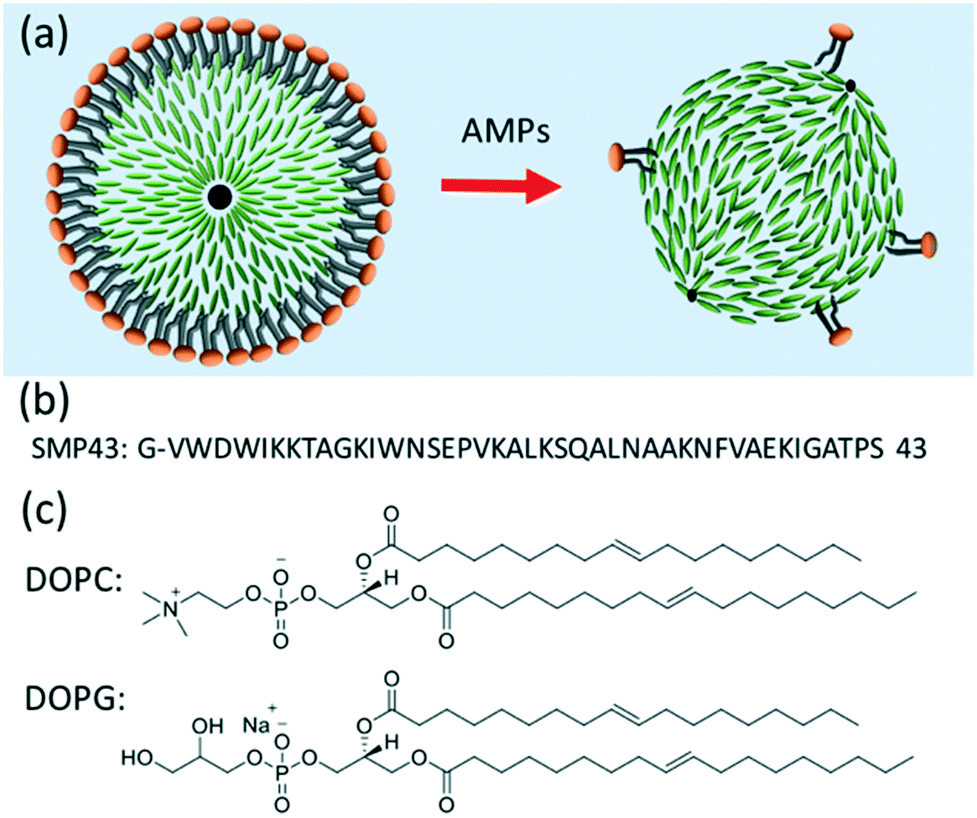
Figure. Schematic diagram showing the bio-sensing mechanism. The lipid coating on the liquid crystal droplet initially induces a radial director geometry. Exposure to the AMP removes the lipid coating, inducing a planar surface alignment and bipolar droplet geometry. LC molecules: green rods. (b) Amino acid sequence of Smp43. (c) Molecular structure of 1,2-dio-leoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-phospho-rac-(1-glycerol) sodium salt (DOPG) lipids used for the monolayer lipid coating on the LC droplet.


