Your basket is currently empty!
ProM1™ MHC Class I Monomers
ProM1™ MHC Class I Monomers, biotin labeled
enabling specialist assays for defined
MHC Class I peptide target interactions
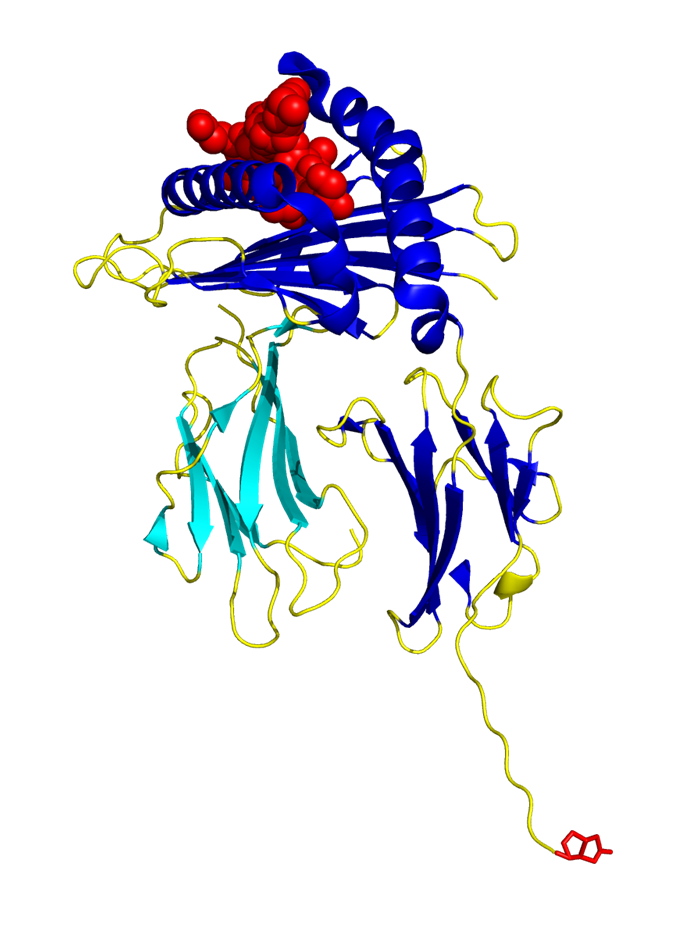
Figure: ProM1™ Monomer
Best in Class MHC Class I Monomers
ProM1™ Class I MHC Monomers are provided already biotin labeled and can be readily adapted to implement specialist solid or solution phase immunoassays or for measuring interactions of predefined MHC Class I-peptide complexes with ligands such as monoclonal antibodies, recombinant T cell receptors, or other binding scaffolds, e.g. for use in screening of novel antigen-specific CAR T cell antigen receptors.
ProM1™ MHC Monomers are made by ProImmune’s experienced team who have been providing superior MHC reagents for more than 18 years now, including our world-leading Pro5® MHC Class I Pentamers.
ProImmune provides ProM1™ Class I MHC Monomers with responsive, expert technical support and customer service. Contrary to ordering class I monomers from in-house or non-commercial facilities, customers do not need to apply and wait for reagents or supply any peptide. ProM1™ Class I MHC Monomers are delivered quickly with stringent quality control and low batch-to-batch variability. As such, ProM1™ Class I MHC Monomers complement ProImmune’s existing Pro5® Class I Pentamer technology for detecting antigen-specific CD8+ T cells.
Features of ProM1™ Class I MHC Monomers
- From the most experienced commercial providers of MHC multimers
- Proven technology for detecting antigen-specific T cells
- Broadest range of catalog items available
- Flexible labelling options
- Long-term storage
- Industry leading customer service from immunologists who understand your work
Available Quantities
35ug, 100ug; please contact us for larger quantities
Catalog ProM1™ Class I MHC Monomers
Catalog ProM1™ Monomers are available within 1-2 weeks of placing an order.
| DISEASE | ALLELE | PEPTIDE CODE | PEPTIDE SEQUENCE | EPITOPE ORIGIN |
|---|---|---|---|---|
| AAV | A*01:01 | 886 | SADNNNSEY | AAV VP1 492-500 |
| Adenovirus | A*01:01 | 686 | TDLGQNLLY | Adenovirus 5 Hexon 886-894 |
| Cancer | A*01:01 | 3623 | DSDPDSFQDY | Tyr A1a 454-463 |
| Cancer | A*01:01 | 3624 | EADPIGHLY | MAGEA3 |
| Cancer | A*01:01 | 29 | EADPTGHSY | MAGE-A1 161-169 |
| Cancer | A*01:01 | 3626 | EVDPASNTY | MAGE-A4 169-177 |
| Cancer | A*01:01 | 30 | EVDPIGHLY | MAGE-A3 168-176 |
| Cancer | A*01:01 | 3627 | HSTNGVTRIY | PSMA |
| Cancer | A*01:01 | 3628 | ILDTAGREEY | N-ras 55-64 |
| Cancer | A*01:01 | 467 | KCDICTDEY | Tyrosinase 243-251 |
| Cancer | A*01:01 | 307 | KSDICTDEY | Tyrosinase 243-251 (244S) |
| Cancer | A*01:01 | 3622 | LTDDRLFTCY | PLEKHM2 |
| Cancer | A*01:01 | 3630 | LVDVMPWLQY | Cytochrome P450 240-249 |
| Cancer | A*01:01 | 1542 | QSLEIISRY | Mcl-1 177-185 |
| Cancer | A*01:01 | 3632 | RSDSGQQARY | AIM-2 |
| Cancer | A*01:01 | 3383 | TLDTLTAFY | Mesothelin 429-437 |
| Cancer | A*01:01 | 3633 | VTEPGTAQY | Minor antigen HA-3T (Lbc oncogene 451-459) |
| Cancer | A*01:01 | 3634 | VYDFFVWLHY | TRP-2 181-190 |
| Cancer | A*01:01 | 2102 | YVDFREYEYY | FLT3 ITD |
| CMV | A*01:01 | 739 | VTEHDTLLY | HCMV pp50 245-253 |
| CMV | A*01:01 | 266 | YSEHPTFTSQY | HCMV pp65 363-373 |
| COVID-19 / SARS-CoV-2 | A*01:01 | 4501 | CTDDNALAYY | SARS-Cov-2 ORF1ab 4163-4172 |
| COVID-19 / SARS-CoV-2 | A*01:01 | 4355 | FTSDYYQLY | SARS-CoV-2 ORF3a 207-215 (confirmed epitope) |
| COVID-19 / SARS-CoV-2 | A*01:01 | 4690 | HTTDPSFLGRY | SARS-CoV-2 ORF1ab 1641-1651 |
| COVID-19 / SARS-CoV-2 | A*01:01 | 4726 | IVDTVSALVY | SARS-CoV-2 ORF1ab 5781-5789 |
| COVID-19 / SARS-CoV-2 | A*01:01 | 4382 | PTDNYITTY | SARS-CoV-2 Replicase polyprotein 1ab 1621-1629 (confirmed epitope) |
| COVID-19 / SARS-CoV-2 | A*01:01 | 4381 | TTDPSFLGRY | SARS-CoV-2 Replicase polyprotein 1ab 1637-1646 (confirmed epitope) |
| HBV | A*01:01 | 3939 | LLDTASALY | Core 30-38 |
| HCV | A*01:01 | 1066 | ATDALMTGF | HCV NS3 1436-1444 |
| HCV | A*01:01 | 740 | ATDALMTGY | HCV NS3 1435-1443 |
| Influenza | A*01:01 | 76 | CTELKLSDY | Influenza A (PR8) NP 44-52 |
| Influenza | A*01:01 | 540 | VSDGGPNLY | Influenza A PB1 591-599 |
| Minor Histocompatibility Antigen | A*01:01 | 652 | IVDCLTEMY | DRRFY 1521-1529 |
| Autoimmunity | A*02:01 | 2259 | ALWGPDPAAA | Proinsulin precursor 15-24 |
| AutoImmunity | A*02:01 | 851 | HLVEALYLV | Insulin B chain 10-18 |
| AutoImmunity | A*02:01 | 151 | KLQVFLIVL | T1D Diabetes human prepro islet amyloid polypeptide ppIAPP 5-13 |
| AutoImmunity | A*02:01 | 954 | LNIDLLWSV | T1D Diabetes IGRP 228-236 |
| Autoimmunity | A*02:01 | 2382 | MVWESGCTV | IA-2 797-805 |
| Autoimmunity | A*02:01 | 969 | RLLPLLALL | Insulin Precursor 76-90 |
| AutoImmunity | A*02:01 | 470 | SLSRFSWGA | Myelin basic protein 110-118 |
| Autoimmunity | A*02:01 | 3394 | VIVMLTPLV | IA-2 805-813 |
| Autoimmunity | A*02:01 | 1904 | VLFGLGFAI | T1D Diabetes IGRP 265-273 |
| AutoImmunity | A*02:01 | 393 | VMNILLQYV | GAD65 114-123 |
| Autoimmunity | A*02:01 | 3342 | YTCPLCRAPV | SSA SS-56 55-64 |
| BK virus | A*02:01 | 1516 | AITEVECFL | VP1 44-52 |
| BK virus | A*02:01 | 3903 | CLLPKMDSV | large T antigen 398-406 |
| BK virus | A*02:01 | 1783 | FLHCIVFNV | large T antigen 410-418 |
| BK virus | A*02:01 | 3781 | LLMWEAVTV | VP1 108-116 |
| Cancer | A*02:01 | 2291 | AILALLPAL | Prostate Stem Cell Antigen (PSCA) 105-133 |
| Cancer | A*02:01 | 3636 | AILALLPALL | PSCA |
| Cancer | A*02:01 | 3420 | AIQDLCLAV | NPM1 |
| Cancer | A*02:01 | 3422 | AIQDLCVAV | NPM1 |
| Cancer | A*02:01 | 543 | ALCNTDSPL | iLR1 |
| Cancer | A*02:01 | 599 | ALDVYNGLL | Prostatic acid phosphatase precursor (PAP) 299-307 |
| Cancer | A*02:01 | 600 | ALFDIESKV | PSM P2 (prostate) |
| Cancer | A*02:01 | 3718 | ALGGHPLLGV | Dickkopf-related protein 1 20-29 |
| Cancer | A*02:01 | 3638 | ALIHHNTHL | HER2 466-474 |
| Cancer | A*02:01 | 550 | ALKDVEERV | MAGE-C2 336-344 |
| Cancer | A*02:01 | 3719 | ALLAGLVSL | FGFR4 676-684 |
| Cancer | A*02:01 | 2860 | ALLPAVPSL | Wilms Tumor Protein 83-91 |
| Cancer | A*02:01 | 1920 | ALLTSRLRFI | Telomerase Reverse Transcriptase (hTRT) 615-624 |
| Cancer | A*02:01 | 3720 | ALLTYMIAHI | Thymidylate synthase 231-240 |
| Cancer | A*02:01 | 3721 | ALMDKSLHV | MART-1 56-64 |
| Cancer | A*02:01 | 1778 | ALMEQQHYV | ITGB8 662-670 |
| Cancer | A*02:01 | 3343 | ALNVYNGLL | ACPP 299-307 |
| Cancer | A*02:01 | 1508 | ALPFGFILV | IL13R 345-353 |
| Cancer | A*02:01 | 3722 | ALPPPLMLL | Heparanase 8-16 |
| Cancer | A*02:01 | 86 | ALQPGTALL | Prostate Stem Cell Antigen (PSCA) 14-22 |
| Cancer | A*02:01 | 3344 | ALSPVPPVV | Bcl-2 85-93 |
| Cancer | A*02:01 | 3723 | ALSVMGVYV | MAGEA9 223-231 |
| Cancer | A*02:01 | 3423 | ALTPVVVTL | cyclin-dependent kinase 4 170-178 |
| Cancer | A*02:01 | 3345 | ALVCYGPGI | FAP alpha 463-471 |
| Cancer | A*02:01 | 3724 | ALVEFEDVL | hnRNP L 140-148 |
| Cancer | A*02:01 | 3725 | ALWPWLLMA | RNF43 11-19 |
| Cancer | A*02:01 | 3346 | ALWPWLLMAT | RNF43 11-20 |
| Cancer | A*02:01 | 3347 | ALYLMELTM | CB9L2 |
| Cancer | A*02:01 | 527 | ALYVDSLFFL | PRAME PRA 300-309 |
| Cancer | A*02:01 | 3726 | AMLGTHTMEV | Melanocyte-specific secreted glycoprotein 184-193 |
| Cancer | A*02:01 | 2569 | AMVGAVLTA | Tyrosinase 482-190 |
| Cancer | A*02:01 | 2582 | ATVGIMIGV | CEACAM5 687-695 |
| Cancer | A*02:01 | 3728 | AVIGALLAV | Melanocyte-specific secreted glycoprotein 20-28 |
| Cancer | A*02:01 | 1130 | AVLPLLELV | MCL-1 139-147 |
| Cancer | A*02:01 | 3350 | CLPSPSTPV | BMI1 271-279 |
| Cancer | A*02:01 | 1779 | CLTSTVQLV | HER-2/neu 789-797 |
| Cancer | A*02:01 | 3730 | CLYGNVEKV | hnRNP L 404-412 |
| Cancer | A*02:01 | 3641 | CMHLLLEAV | MG50 624-632 |
| Cancer | A*02:01 | 3731 | DLIFGLNAL | Heparanase 185-193 |
| Cancer | A*02:01 | 82 | ELAGIGILTV | MelanA / MART 26-35 |
| Cancer | A*02:01 | 3732 | ELFQDLSQL | ETV5 54-53 |
| Cancer | A*02:01 | 3352 | ELSDSLGPV | PASD1 695-703 |
| Cancer | A*02:01 | 3733 | FAWERVRGL | Cyclin-dependent kinase inhibitor 1 97-105 |
| Cancer | A*02:01 | 3734 | FIASNGVKLV | ACTN4 118-127 (K5N) |
| Cancer | A*02:01 | 2083 | FLAEDALNTV | Epithelial Discoidin Domain Receptor 1 (EDDR1) 867-876 |
| Cancer | A*02:01 | 1302 | FLAKLNNTV | HCA587 317-325 |
| Cancer | A*02:01 | 3735 | FLALIICNA | Tubulin beta 4 283-291 |
| Cancer | A*02:01 | 3736 | FLDEFMEGV | Malic enzyme 224-232 |
| Cancer | A*02:01 | 2282 | FLDPRPLTV | CYP190 |
| Cancer | A*02:01 | 3355 | FLFLRNFSL | TARP(V28L)27-35 |
| Cancer | A*02:01 | 1013 | FLGYLILGV | Prostatic Acid Phosphatase-3 (PAP-3) |
| Cancer | A*02:01 | 3643 | FLIIWQNTM | FSP26 |
| Cancer | A*02:01 | 3356 | FLPSPLFFFL | TARP(P5L) 5-13 |
| Cancer | A*02:01 | 3644 | FLPWHRLFLL | Tyrosinase 207-216 |
| Cancer | A*02:01 | 998 | FLTGNQLAV | 5T4 97-105 |
| Cancer | A*02:01 | 404 | FLTPKKLQCV | Prostate Specific Antigen-1 (PSA-1) 141-150 |
| Cancer | A*02:01 | 34 | FLWGPRALV | MAGEA3 271-279 |
| Cancer | A*02:01 | 2079 | FLYDDNQRV | Topoisomerase II-alpha-b 828-836 |
| Cancer | A*02:01 | 306 | FLYTLLREV | STEAP 86-94 |
| Cancer | A*02:01 | 163 | FMNKFIYEI | Human alfa fetoprotein 158-166 |
| Cancer | A*02:01 | 813 | FVGEFFTDV | GPC3 144-152 (overexpressed in hepatocellular carcinoma) |
| Cancer | A*02:01 | 3647 | FVWLHYYSV | TRP2 185-193(L) |
| Cancer | A*02:01 | 1386 | GLAPPQHLIRV | p53 187-197 |
| Cancer | A*02:01 | 3648 | GLFGDIYLA | CSNK1A1 26-34 |
| Cancer | A*02:01 | 3649 | GLFGDIYLAI | CSNK1A1 26-35 |
| Cancer | A*02:01 | 3357 | GLFKCGIAV | FAP 639-647 |
| Cancer | A*02:01 | 3358 | GLIQLVEGV | TRAG-3 4-12 |
| Cancer | A*02:01 | 1786 | GLLGASVLGL | Telomerase Reverse Transcriptase (hTRT) 674-683 |
| Cancer | A*02:01 | 560 | GLMEEMSAL | Human Mena protein (overexpressed in breast cancer) |
| Cancer | A*02:01 | 2236 | GLMKYIGEV | TRPM8 187-195 |
| Cancer | A*02:01 | 2293 | GLQHWVPEL | BA46 (Lactadherin) 97-106 |
| Cancer | A*02:01 | 2778 | GLSNLTHVL | PRAME (432-440) |
| Cancer | A*02:01 | 164 | GLSPNLNRFL | Alpha-fetoprotein isoform 2 167-176 |
| Cancer | A*02:01 | 389 | GLYDGMEHL | MAGEA-10 254-262 |
| Cancer | A*02:01 | 2669 | GVLLWEIFSL | VEGFR1 28-37 |
| Cancer | A*02:01 | 125 | GVLVGVALI | Carcinogenic Embryonic Antigen (CEA) 694-702 |
| Cancer | A*02:01 | 2294 | GVRGRVEEI | BCR-ABL |
| Cancer | A*02:01 | 575 | GVYDGREHTV | MAGE-A4 230-239 |
| Cancer | A*02:01 | 249 | HLSTAFARV | G250 (renal cell carcinoma) 217-225 |
| Cancer | A*02:01 | 226 | HLYQGCQVV | Receptor tyrosine-protein kinase erbB-2 48-56 |
| Cancer | A*02:01 | 85 | ILAKFLHWL | Telomerase 540-548 |
| Cancer | A*02:01 | 3361 | ILGVLTSLV | DLK1 309-317 |
| Cancer | A*02:01 | 811 | ILHDGAYSL | HER-2 434-443 |
| Cancer | A*02:01 | 210 | ILHNGAYSL | HER-2/neu 435-443 |
| Cancer | A*02:01 | 3654 | ILLRDAGLV | TRAG-3L 58-66 |
| Cancer | A*02:01 | 3655 | ILLVVVLGV | Receptor tyrosine-protein kinase erbB-2 707-715 |
| Cancer | A*02:01 | 329 | ILLWQPIPV | Prostatic Acid Phosphatase-3 (PAP-3) 135-143 |
| Cancer | A*02:01 | 3656 | ILNAMIAKI | HAUS3 154-162 |
| Cancer | A*02:01 | 468 | ILSLELMKL | Receptor for hyaluronic acid-mediated motility (RHAMM) 165-173 |
| Cancer | A*02:01 | 46 | IMDQVPFSV | gp100 (pmel17) 209-217 |
| Cancer | A*02:01 | 2296 | ITDQVPFSV | gp100 (pmel) 209-217 |
| Cancer | A*02:01 | 1246 | KASEKIFYV | SSX2 41-49 |
| Cancer | A*02:01 | 3658 | KASEYLQLV | MAGEA2 153-161 |
| Cancer | A*02:01 | 214 | KIFGSLAFL | HER-2/neu 369-377 |
| Cancer | A*02:01 | 3659 | KIWEELSVL | MAGEA3 220-228 |
| Cancer | A*02:01 | 2297 | KLCPVQLWV | p53 139-147 |
| Cancer | A*02:01 | 646 | KLFGTSGQKT | EGF-R-479 350-359 |
| Cancer | A*02:01 | 3660 | KLIDRTE(S)L | LSP1 325-333 |
| Cancer | A*02:01 | 3416 | KLMSSNSTDL | HSP105 234-243 |
| Cancer | A*02:01 | 405 | KLQCVDLHV | Prostate Specific Antigen 146-154 |
| Cancer | A*02:01 | 859 | KLQDASAEV | HM1.24-aa 126-134 |
| Cancer | A*02:01 | 3661 | KLTGDENFTI | Tyrosinase precursor 224-233 |
| Cancer | A*02:01 | 655 | KLVVVGADGV | KRAS G12D (5-14) |
| Cancer | A*02:01 | 752 | KLVVVGAVGV | KRAS G12V (5-14) |
| Cancer | A*02:01 | 57 | KTWGQYWQV | gp100 (pmel17) 154-162 |
| Cancer | A*02:01 | 2298 | KVAEELVHFL | MAGEA3 112-120 (alternative version) |
| Cancer | A*02:01 | 302 | KVAELVHFL | MAGEA3 112-120 |
| Cancer | A*02:01 | 3006 | KVDELAHFL | Melanoma-associated antigen 4 (MAGE4) 113-121 |
| Cancer | A*02:01 | 3080 | KVLEHVVRV | Melanoma-associated antigen 4 (MAGE4) 286-294 |
| Cancer | A*02:01 | 171 | KVLEYVIKV | MAGEA1 278-286 |
| Cancer | A*02:01 | 1421 | KVVEFLAML | MAGE-C1 1083-1091 |
| Cancer | A*02:01 | 810 | LIAHNQVRQV | HER-2/neu (85-94) |
| Cancer | A*02:01 | 628 | LLAARAIVAI | iLR1 59-68 |
| Cancer | A*02:01 | 3662 | LLCYSCKAQV | PSCA 17-26 |
| Cancer | A*02:01 | 221 | LLDVAPLSL | HSP1A 461-469 |
| Cancer | A*02:01 | 1303 | LLFGLALIEV | MAGE-C2 191-200 |
| Cancer | A*02:01 | 516 | LLGATCMFV | CyclinD 101-109 |
| Cancer | A*02:01 | 205 | LLGRNSFEV | p53 264-272 |
| Cancer | A*02:01 | 690 | LLHETDSAV | PSMA/PSM-P1 4-12 |
| Cancer | A*02:01 | 1007 | LLLAGLFSL | Fibromodulin 7-15 |
| Cancer | A*02:01 | 3666 | LLLEAVPAV | MG50 69-77 |
| Cancer | A*02:01 | 841 | LLLIWFRPV | BKV Ltag 579-587 |
| Cancer | A*02:01 | 220 | LLLLDVAPL | HSP1A 459-467 |
| Cancer | A*02:01 | 208 | LLLLTVLTV | MUC-1 12-20 |
| Cancer | A*02:01 | 381 | LLLTVLTVV | Tumor Mucin Antigen 13-21 |
| Cancer | A*02:01 | 3668 | LLNQLQVNL | Mucin2 467-475 |
| Cancer | A*02:01 | 3669 | LLRDAGLVKM | TRAP 59-68 |
| Cancer | A*02:01 | 3670 | LLRRYNVAKV | SOX11 266-275 |
| Cancer | A*02:01 | 3671 | LLSHGAVIEV | Ankyrin NYBR1 158-167 |
| Cancer | A*02:01 | 3363 | LLVPTCVFLV | 691-700 |
| Cancer | A*02:01 | 2793 | LMAQEALAFL | CAMEL 2-11 |
| Cancer | A*02:01 | 391 | LMLGEFLKL | Survivin 96-104 |
| Cancer | A*02:01 | 1464 | LTLGEFLKL | Survivin-3A 96-104 |
| Cancer | A*02:01 | 3673 | LVFGIELMEV | MAGEA3 160-169 |
| Cancer | A*02:01 | 3674 | LVFGIEVVEV | MAGEA12 160-169 |
| Cancer | A*02:01 | 3365 | MLAVFLPIV | STEAP 292-300 (293L) |
| Cancer | A*02:01 | 3675 | MLWGWREHV | Mucin2 645-653 |
| Cancer | A*02:01 | 3366 | NLFETPVEA | 194-202 |
| Cancer | A*02:01 | 480 | PLFDFSWLSL | Bcl-2 208-217 |
| Cancer | A*02:01 | 162 | PLFQVPEPV | Alpha-fetoprotein isoform 1 137-145 |
| Cancer | A*02:01 | 3678 | PLQPEQLQV | Receptor tyrosine-protein kinase erbB-2 437-445 |
| Cancer | A*02:01 | 662 | PLTSIISAV | Receptor tyrosine-protein kinase erbB-2 728-736 |
| Cancer | A*02:01 | 338 | QLCPICRAPV | Livin/ML-IAP280 175-184 |
| Cancer | A*02:01 | 780 | QLFEELQEL | Heme oxygenase-1 212-220 |
| Cancer | A*02:01 | 3368 | QLGEQCWTV | PSCA 44-51 (51A) |
| Cancer | A*02:01 | 1908 | QLLDGFMITL | PASD1 39-48 |
| Cancer | A*02:01 | 3421 | QLLIKAVNL | MPP11 |
| Cancer | A*02:01 | 3680 | QLMAFNHLI | PAX3/FKHR 135-143 |
| Cancer | A*02:01 | 3681 | QLMPYGCLL | Receptor tyrosine-protein kinase erbB-2 845-853 |
| Cancer | A*02:01 | 3371 | RLAEYQAYI | SART3 309-317 |
| Cancer | A*02:01 | 997 | RLARLALVL | Trophoblast glycoprotein 17-25 |
| Cancer | A*02:01 | 787 | RLASFYDWPL | BIR7 90-99 |
| Cancer | A*02:01 | 3684 | RLGPTLMCL | MG50 1244-1252 |
| Cancer | A*02:01 | 212 | RLLQETELV | HER-2/neu 689-697 |
| Cancer | A*02:01 | 960 | RLMNDMTAV | HSP105 128-136 |
| Cancer | A*02:01 | 337 | RLQEERTCKV | BIR |
| Cancer | A*02:01 | 3436 | RLQGISPKI | SSX2 103-111 |
| Cancer | A*02:01 | 1937 | RLSSCVPVA | 131-139 |
| Cancer | A*02:01 | 3687 | RLTRFLSRV | CyclinD 228-236 |
| Cancer | A*02:01 | 117 | RLVDDFLLV | Telomerase Reverse Transcriptase 865-873 |
| Cancer | A*02:01 | 884 | RLWQELSDI | circadian clock protein PASD1 691-700 |
| Cancer | A*02:01 | 202A | RMFPNAPYL | WT-1 126-134 (Wilms tumor) |
| Cancer | A*02:01 | 654 | RMPEAAPPV | p53 65-73 |
| Cancer | A*02:01 | 3688 | RTF(S)PTYGL | Desmuslin 426-434 |
| Cancer | A*02:01 | 2988 | RVA(PHOSPHO-S)PTSGV | Insulin receptor substrate-2 1097-1105 |
| Cancer | A*02:01 | 2990 | RVASPTSGV | IRS-2 1097-1105 |
| Cancer | A*02:01 | 3374 | SIDWFMVTV | p31-39 |
| Cancer | A*02:01 | 3689 | SILLRDAGL | TRAP 57-65 |
| Cancer | A*02:01 | 3375 | SILLRDAGLV | TRAG-3 57-66 |
| Cancer | A*02:01 | 3691 | SLADEAEVYL | GAS7 Neoepitope |
| Cancer | A*02:01 | 637 | SLAMLDLLHV | Mutant anaplastic lymphoma kinase 1220-1229 |
| Cancer | A*02:01 | 3693 | SLDDYNHLV | L-dopachrome tautomerase 288-296 |
| Cancer | A*02:01 | 457 | SLEENIVIL | RHAMM 275-283 |
| Cancer | A*02:01 | 3377 | SLFEPPPPG | PSMA 85-93 |
| Cancer | A*02:01 | 1056 | SLFLGILSV | CD20 188-196 (B cell malignancies) |
| Cancer | A*02:01 | 452 | SLGEQQYSV | WT1 187-195 |
| Cancer | A*02:01 | 774 | SLLFLLFSL | Mesothelin 20-28 |
| Cancer | A*02:01 | 49 | SLLMWITQC | NY-ESO-1 157-165 (9C) |
| Cancer | A*02:01 | 390 | SLLMWITQV | NY-ESO-1 157-165 |
| Cancer | A*02:01 | 861 | SLLQHLIGL | PRAME 425-433 |
| Cancer | A*02:01 | 2299 | SLPPPGTRV | p53 149-157 |
| Cancer | A*02:01 | 1181 | SLSEKTVLL | CD59 glycoprotein precursor 106-114 |
| Cancer | A*02:01 | 958 | SLVDVMPWL | Cytochrome p450 1B1 239-248 |
| Cancer | A*02:01 | 3695 | SLYKFSPFPL | O-linked N-acetylglucosamine transferase FSP06 |
| Cancer | A*02:01 | 3417 | SLYSYFQKV | Membrane-associated transporter protein |
| Cancer | A*02:01 | 3696 | SMTR(S)PPRV | SFRS2B 241-249 |
| Cancer | A*02:01 | 3379 | SQADALKYV | EZH2 729-737 |
| Cancer | A*02:01 | 3418 | STLCQVEPV | MPP11 |
| Cancer | A*02:01 | 1509 | TLADFDPRV | EphA2 |
| Cancer | A*02:01 | 408 | TLAPATEPA | Mucin 79-87 |
| Cancer | A*02:01 | 3698 | TLEEITGYL | Receptor tyrosine-protein kinase erbB-2 448-456 |
| Cancer | A*02:01 | 3699 | TLHCDCEIL | MG50 210-218 |
| Cancer | A*02:01 | 915 | TLPGYPPHV | PAX-5 311-319 |
| Cancer | A*02:01 | 2371 | TLQDIVYKL | BMI1 74-82 |
| Cancer | A*02:01 | 561 | TMNGSKSPV | hMena 502-510 |
| Cancer | A*02:01 | 862 | VIFDFLHCI | BKV Ltag 406-414 |
| Cancer | A*02:01 | 1403 | VIMPCSWWV | Chondromodulin-I 319-327 |
| Cancer | A*02:01 | 97 | VISNDVCAQV | Prostate Specific Antigen-1 (PSA-1) 154-163 |
| Cancer | A*02:01 | 689 | VLAGGFFLL | PSMA 27-38 |
| Cancer | A*02:01 | 304 | VLAGVGFFI | EPHA2 550-558 |
| Cancer | A*02:01 | 893 | VLDFAPPGA | WT1 |
| Cancer | A*02:01 | 781 | VLDGLDVLL | PRAME 100-108 |
| Cancer | A*02:01 | 3702 | VLEPPGARDV | BIR 7 230-239 |
| Cancer | A*02:01 | 345 | VLGEAWRDQV | TRAP 45-54 |
| Cancer | A*02:01 | 3704 | VLLALLMAGL | Prostate stem cell antigen 4-13 |
| Cancer | A*02:01 | 773 | VLPLTVAEV | Mesothelin 530-538 |
| Cancer | A*02:01 | 250 | VLQELNVTV | Leukocyte Proteinase-3 (Wegener's autoantigen) 169-177 |
| Cancer | A*02:01 | 3419 | VLQMKEEDV | iLR1 |
| Cancer | A*02:01 | 3705 | VLSVNVPDV | MG50 625-633 |
| Cancer | A*02:01 | 3706 | VLVKSPNHV | Receptor tyrosine-protein kinase erbB-4 890-898 |
| Cancer | A*02:01 | 157 | VLYRYGSFSV | gp100 (pmel17) 476-485 |
| Cancer | A*02:01 | 693 | VMAGVGSPYV | Receptor tyrosine-protein kinase erbB-2 819-828 |
| Cancer | A*02:01 | 3708 | VMIG(S)PKKV | Tensin3 1558-1566 |
| Cancer | A*02:01 | 3709 | VVLGVVFGI | Receptor tyrosine-protein kinase erbB-2 743-751 |
| Cancer | A*02:01 | 3710 | WLPKILGEV | MG50 1051-1059 |
| Cancer | A*02:01 | 3711 | WLQYFPNPV | Cytochrome P450 246-254 |
| Cancer | A*02:01 | 3386 | WLSLKTLLSL | Bcl-2 214-223 |
| Cancer | A*02:01 | 2457 | YAIDLPVSV | L-dopachrome tautomerase 488-496 |
| Cancer | A*02:01 | 58 | YLEPGPVTA | gp100 |
| Cancer | A*02:01 | 47 | YLEPGPVTV | gp100 (pmel) 280-288 (288V) |
| Cancer | A*02:01 | 394 | YLFFYRKSV | mTERT 572-580 |
| Cancer | A*02:01 | 2300 | YLGSYGFRL | p53 103-111 |
| Cancer | A*02:01 | 2081 | YLIELIDRV | TACE 250-258 |
| Cancer | A*02:01 | 1239 | YLISGDSPV | CD33 65-73 (1Y2L) |
| Cancer | A*02:01 | 3713 | YLLDLSTNHL | Fibromodulin 7-15 |
| Cancer | A*02:01 | 594 | YLNDHLEPWI | Bcl-X 173-182 |
| Cancer | A*02:01 | 2194 | YLNPSVDSV | EWS-FLI-1 oncogene |
| Cancer | A*02:01 | 3387 | YLNRHLHTWI | BCL-2 180-189 |
| Cancer | A*02:01 | 645 | YLNTVQPTCV | EGF-R 1138-1147 |
| Cancer | A*02:01 | 922 | YLQHNEIQEV | Fibromodulin 206-215 |
| Cancer | A*02:01 | 2301 | YLQLVFGIEV | MAGEA2 157-166 |
| Cancer | A*02:01 | 447 | YLQVNSLQTV | Telomerase Reverse Transcriptase (hTRT) 988-997 |
| Cancer | A*02:01 | 3388 | YLQWIEFSI | Prominin1 744-752 |
| Cancer | A*02:01 | 48 | YLSGADLNL | Carcinoembryonic antigen (CEA)-derived peptide CAP1-6D |
| Cancer | A*02:01 | 75 | YLSGANLNL | Carcinogenic Embryonic Antigen (CEA) 571-579 |
| Cancer | A*02:01 | 1910 | YLVGNVCIL | PASD1 168-176 |
| Cancer | A*02:01 | 3715 | YLWWVNNQSL | CEA 176-185 |
| Cancer | A*02:01 | 3389 | YLYQWLGAPV | Osteocalcin 51-60 |
| Cancer | A*02:01 | 1201 | YMCSFLFNL | Ewing Tumor EZH2 666-674 |
| Cancer | A*02:01 | 61 | YMDGTMSQV | Tyrosinase 369-377 (371D) |
| Cancer | A*02:01 | 921 | YMEHNNVYTV | Fibromodulin 250-259 |
| Cancer | A*02:01 | 464 | YMNGTMSQV | Tyrosinase 368-376 |
| Cancer | A*02:01 | 2649 | YVDPVITSI | Hepatocyte growth factor receptor 673-681 |
| Chlamydia | A*02:01 | 335 | NMFTPYIGV | MOMP precursor 283-291 |
| Chlamydia | A*02:01 | 334 | RLNMFTPYI | Chlamydia trachomatis MOMP 258-266 |
| CMV | A*02:01 | 2205 | MLNIPSINV | pp65 120-128 |
| CMV | A*02:01 | 8 | NLVPMVATV | HCMV pp65 495-504 |
| CMV | A*02:01 | 1598 | VLAELVKQI | HCMV IE1 81-89 |
| CMV | A*02:01 | 144 | VLEETSVML | HCMV IE1 316-324 (UL123) |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4322 | ALNTLVKQL | SARS-CoV Spike glycoprotein precursor 940-948 (conserved in SARS-CoV-2) |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4321 | FIAGLIAIV | SARS-CoV-2 Spike glycoprotein 1220-1228 (confirmed epitope) |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4340 | KIADYNYKL | SARS-CoV-2 Spike glycoprotein 417-425 |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4418 | KLWAQCVQL | SARS-CoV-2 ORF1ab 3886-3894 (confirmed epitope) |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4323 | LITGRLQSL | SARS-CoV-2 Spike glycoprotein 996-1004 (confirmed epitope) |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4320 | LLLDRLNQL | SARS-CoV Nucleocapsid protein 223-231 (conserved in SARS-CoV-2) |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4342 | LLYDANYFL | SARS-CoV-2 ORF3a 139-147 (confirmed epitope) |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4324 | NLNESLIDL | SARS-CoV Spike glycoprotein precursor 1174-1182 (conserved in SARS-Cov-2) |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4358 | RLQSLQTYV | SARS-CoV-2 Spike glycoprotein 1000-1008 (confirmed subdominant epitope) |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4725 | TLVPQEHYV | SARS-CoV-2 ORF1ab 5563-5571 |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4326 | VLNDILSRL | SARS-CoV Spike glycoprotein precursor 958-966 (conserved in SARS-Cov-2) |
| COVID-19 / SARS-CoV-2 | A*02:01 | 4339 | YLQPRTFLL | SARS-Cov-2 Spike glycoprotein 269-277 (confirmed epitope) |
| Dengue | A*02:01 | 3918 | AIKRGLRTL | Dengue NS3 112-120 |
| Dengue | A*02:01 | 3914 | ILIRTGLLVI | Dengue NS2b 97-106 |
| Dengue | A*02:01 | 3931 | KLAEAIFKL | Dengue NS5 563-571 |
| Dengue | A*02:01 | 3923 | LLLGLMILL | Dengue NS4a 56-64 |
| Dengue | A*02:01 | 2929 | TLYAVATTI | Dengue NS4b 40-48 |
| Dengue | A*02:01 | 3928 | VLLLVTHYA | Dengue NS4b 111-119 |
| EBV | A*02:01 | 42 | CLGGLLTMV | EBV LMP-2 426-434 |
| EBV | A*02:01 | 2292 | FLDKGTYTL | EBV BALF-4 276-284 |
| EBV | A*02:01 | 666 | FLYALALLL | EBV LMP-2 356-364 |
| EBV | A*02:01 | 3462 | FMVFLQTHI | EBV EBNA-1 (562-570) |
| EBV | A*02:01 | 1 | GLCTLVAML | EBV BMLF-1 259-267 |
| EBV | A*02:01 | 1616 | LLDFVRFMGV | EBV EBNA-3C 284-293 |
| EBV | A*02:01 | 872 | TLDYKPLSV | EBV BMRF1 208-216 |
| EBV | A*02:01 | 409 | YLLEMLWRL | EBV LMP-1 125-133 |
| EBV | A*02:01 | 410 | YLQQNWWTL | EBV LMP1 159-167 |
| EBV | A*02:01 | 462 | YVLDHLIVV | EBV BRLF1 109-117 |
| HAV | A*02:01 | 2257 | LLYNCCYHV | Hepatitis A Virus polyprotein 2027-2034 |
| HBV | A*02:01 | 3934 | ELMTLATWV | core protein 64-72 |
| HBV | A*02:01 | 32 | FLLSLGIHL | HBV polymerase 573-581 |
| HBV | A*02:01 | 27 | FLLTRILTI | HBV envelope 183-191 |
| HBV | A*02:01 | 283 | FLPSDFFPSI | HBV core 18-27 (subtype ADR4) |
| HBV | A*02:01 | 23 | FLPSDFFPSV | HBV core antigen 18-27 |
| HBV | A*02:01 | 28 | GLSPTVWLSV | HBV surface antigen 185-194 |
| HBV | A*02:01 | 778 | GLSRYVARL | HBV Pol 455-463 |
| HBV | A*02:01 | 2829 | KLHLYSHPI | Pol 502-510 |
| HBV | A*02:01 | 500 | VLHKRTLGL | X 92-100 |
| HBV | A*02:01 | 31 | WLSLLVPFV | HBV surface antigen 172-181 |
| HBV | A*02:01 | 1194 | YMDDVVLGA | Polymerase 548-556 |
| HCV | A*02:01 | 37 | ALYDVVTKL | HCV NS5b 2594-2602 |
| HCV | A*02:01 | 3 | CINGVCWTV | HCV NS3 1073-1081 |
| HCV | A*02:01 | 858 | CVNGVCWTV | HCV NS3 1073-1081 |
| HCV | A*02:01 | 2 | DLMGYIPAV | HCV core 132-140 |
| HCV | A*02:01 | 491 | DLMGYIPLV | HCV core 132-140 |
| HCV | A*02:01 | 1094 | GLQDCTMLV | HCV NS5B 2727-2735 |
| HCV | A*02:01 | 242 | KLSGLGINAV | HCV NS3 1406-1415 |
| HCV | A*02:01 | 243 | KLSGLGLNAV | HCV NS3 1406-1415 |
| HCV | A*02:01 | 40 | KLVALGINAV | HCV NS3 1406-1415 |
| HCV | A*02:01 | 172 | LLFNILGGWV | HCV NS4b 1807-1816 |
| HCV | A*02:01 | 36 | VLSDFKTWL | HCV NS5a 1987-1995 |
| HCV | A*02:01 | 4 | YLLPRRGPRL | HCV core 35-44 |
| HIV/SIV | A*02:01 | 1240 | ALVEMGHHA | HIV Vpu 66-74 |
| HIV/SIV | A*02:01 | 1147 | FLGKIWPS | Gag 433-440 |
| HIV/SIV | A*02:01 | 430 | GLADQLIHL | HIV-1 Vif 101-109 |
| HIV/SIV | A*02:01 | 84 | ILKEPVHGV | HIV-1 RT 476-484 |
| HIV/SIV | A*02:01 | 246 | KLTPLCVTL | HIV-1 env gp120 90-98 |
| HIV/SIV | A*02:01 | 799 | LTFGWCFKL | HIV-1 Nef 137-145 |
| HIV/SIV | A*02:01 | 1244 | NVWATHACV | HIV Env gp 67-7 |
| HIV/SIV | A*02:01 | 1241 | RTLNAWVKV | HIV Gag 150-158 |
| HIV/SIV | A*02:01 | 3401 | SLFNTVATL | HIV gag 77-85 |
| HIV/SIV | A*02:01 | 1537 | SLLNATAIAV | HIV env 816-825 |
| HIV/SIV | A*02:01 | 3402 | SLVKHHMYI | HIV Vif 23-31 |
| HIV/SIV | A*02:01 | 10 | SLYNTVATL | HIV-1 gag p17 76-84 |
| HIV/SIV | A*02:01 | 174 | TLNAWVKVV | HIV-1 gag p24 19-27 |
| HIV/SIV | A*02:01 | 3403 | VIYHYVDDL | HIV Pol |
| HPV | A*02:01 | 658 | KLPQLCTEL | HPV 16 E6 18-26 |
| HPV | A*02:01 | 2312 | MLDLQPETT | HPV 16 E7 12-20 |
| HPV | A*02:01 | 2103 | YMLDLQPET | HPV E7 11-19 |
| HPV | A*02:01 | 95 | YMLDLQPETT | HPV 16 E7 11-20 |
| HSV | A*02:01 | 1153 | QLFNHTMFI | Non-muscle Myosin 478-486 |
| HSV | A*02:01 | 3406 | QMARLAWEA | 1116-1124 |
| HSV | A*02:01 | 1152 | VLMIKALEL | Non muscle Myosin-9 741-749 |
| HTLV | A*02:01 | 2309 | AVLDGLLSL | HTLV bZIP factor 42-50 |
| HTLV | A*02:01 | 3408 | GLLSLEEEL | bZIP factor 26-34 |
| HTLV | A*02:01 | 200 | LLFGYPVYV | Human T-cell lymphotropic virus-1 (HTLV-1) tax 11-19 |
| Influenza | A*02:01 | 7 | GILGFVFTL | Influenza A MP 58-66 |
| Influenza | A*02:01 | 2295 | ILGFVFTLTV | Influenza A MP 59-68 |
| Influenza | A*02:01 | 3409 | KLGEFYNQMM | Flu BNP 85-94 (Influenza B) |
| JC Virus | A*02:01 | 611 | ILMWEAVTL | VP1 100-108 |
| JC Virus | A*02:01 | 610 | SITEVECFL | VP1 36-44 |
| LCMV | A*02:01 | 2308 | ALPHIIDEV | LCMV envelope gp 10-18 |
| LCMV | A*02:01 | 3411 | SLNQTVHSL | NP 69-77 |
| LCMV | A*02:01 | 2310 | YLVSIFLHL | LCMV envelope gp 447-455 |
| Malaria | A*02:01 | 314 | YLNKIQNSL | Plasmodium falciparum CSP 334-342 |
| Minor Histocompatibility Antigen | A*02:01 | 175 | FIDSYICQV | miHAg H-Y (human SMCY) 311-319 |
| Minor Histocompatibility Antigen | A*02:01 | 581 | RTLDKVLEV | miHAg HA-8 |
| Minor Histocompatibility Antigen | A*02:01 | 473 | VLHDDLLEA | Minor Histocompatibility Antigen HA-1 137-145 |
| Minor Histocompatibility Antigen | A*02:01 | 395 | YIGEVLVSV | HA-2 |
| Negative | A*02:01 | N01 | NEGATIVE | Negative Control |
| Norovirus | A*02:01 | 3214 | TMFPHIIVDV | Norovirus VP1 139-148 |
| Other | A*02:01 | 3041 | ILEDIVLTL | Streptococcus pyogenes Cas9 615-623 |
| Other | A*02:01 | 133 | LLDVPTAAV | Interferon gamma inducible protein (GILT) 30 27-35 |
| Other | A*02:01 | 366 | LMWYELSKI | KSHVF-8 gB.492-500 |
| Other | A*02:01 | 353 | RILGAVAKV | Vinculin 822-830 |
| Other | A*02:01 | 370 | TIHDIILECV | Alpha papillomavirus 9 E6 Protein (22-31) |
| RSV | A*02:01 | 801 | KMLKEMGEV | RSV NP 137-145 |
| Tuberculosis | A*02:01 | 620 | AMASTEGNV | ESAT-6 |
| Tuberculosis | A*02:01 | 619 | GILTVSVAV | 16 kDa |
| Tuberculosis | A*02:01 | 324 | GLPVEYLQV | Mycobacterium bovis antigen 85-A 6-14 |
| Tuberculosis | A*02:01 | 276 | KLIANNTRV | Mycobacterium bovis antigen 85-A 200-208 |
| Tuberculosis | A*02:01 | 2221 | VLTDGNPPEV | 19 kDa |
| Vaccinia | A*02:01 | 978 | ILDDNLYKV | Vaccinia virus Copenhagen Protein G5 18-26 |
| Vaccinia | A*02:01 | 703 | KVDDTFYYV | Vaccinia virus Host range protein 2 74-82 |
| Varicella Zoster Virus | A*02:01 | 995 | ALWALPHAA | IE62 593-601 |
| West Nile | A*02:01 | 1322 | ATWAENIQV | West Nile virus NY-99 polyprotein precursor 3390-3398 |
| West Nile | A*02:01 | 1320 | RLDDDGNFQL | West Nile Virus NY-99 polyprotein precursor (1452-1461) |
| West Nile | A*02:01 | 1327 | YTMDGEYRL | West Nile virus NY-99 polyprotein precursor 2023-2031 |
| West Nile Virus | A*02:01 | 1324 | SLFGQRIEV | WNV nonstructural protein 4B 15-23 |
| West Nile Virus | A*02:01 | 1321 | SVGGVFTSV | WNV envelope gp 430-438 |
| Yellow Fever Virus | A*02:01 | 2097 | LLWNGPMAV | NS4B 214-222 |
| Cancer | A*03:01 | 118 | ALLAVGATK | gp100 (pmel17) 17-25 |
| Cancer | A*03:01 | 264 | ATGFKQSSK | bcr-abl 210 kD fusion protein 259-269 |
| Cancer | A*03:01 | 87 | KQSSKALQR | bcr-abl 210 kD fusion protein 21-29 |
| Cancer | A*03:01 | 3369 | QVLKKIAQK | HMOX1 145-153 |
| Cancer | A*03:01 | 3370 | RIAAWMATY | 165-173 |
| Cancer | A*03:01 | 541 | RISTFKNWPK | Survivin-3A 18-27 (27K) |
| Cancer | A*03:01 | 552 | RLGLQVRKNK | RhoC 176-185 (177L) |
| Cancer | A*03:01 | 615 | RLLFFAPTR | Mcl-1 95-103 |
| CMV | A*03:01 | 1599 | KLGGALQAK | HCMV IE1 184-192 |
| COVID-19 / SARS-CoV-2 | A*03:01 | 4443 | KCYGVSPTK | SARS-CoV-2 Spike glycoprotein 378-386 |
| COVID-19 / SARS-CoV-2 | A*03:01 | 4724 | KLFAAETLK | SARS-CoV-2 ORF1ab 5455-5463 |
| COVID-19 / SARS-CoV-2 | A*03:01 | 4356A | KTFPPTEPK | SARS-CoV-2 Nucleocapsid protein 362-370 (confirmed epitope) |
| COVID-19 / SARS-CoV-2 | A*03:01 | 4679A | SVYAWNRKR | SARS-CoV-2 Spike glycoprotein 349-357 |
| COVID-19 / SARS-CoV-2 | A*03:01 | 4691A | VTNNTFTLK | SARS-CoV-2 ORF1ab 808-816 |
| Dengue | A*03:01 | 3913 | ELERAADVK | Dengue NS2b 52-60 |
| Dengue | A*03:01 | 3929A | KITAEWLWK | Dengue NS5 375-383 |
| Dengue | A*03:01 | 3916A | RIEPSWADVK | Dengue NS3 64-74 |
| Dengue | A*03:01 | 3921A | RVIDPRRCMK | Dengue NS3 422-431 |
| Dengue | A*03:01 | 3910A | RVSTVQQLTK | Dengue C 22-31 |
| EBV | A*03:01 | 727 | RLRAEAQVK | EBV EBNA 3A 603-611 |
| EBV | A*03:01 | 1838 | RVRAYTYSK | EBV BRLF1 |
| HCV | A*03:01 | 35 | RVCEKMALY | HCV NS5B 2588-2596 |
| HIV/SIV | A*03:01 | 1496 | AIFQSSMTK | HIV pol 325-333 |
| HIV/SIV | A*03:01 | 109 | QVPLRPMTYK | HIV-1 nef 73-82 |
| HIV/SIV | A*03:01 | 194 | RLRPGGKKK | HIV-1 gag p17 19-27 |
| HPV | A*03:01 | 2064 | KLCLRFLSK | HPV 33 E6 64-72 |
| Influenza | A*03:01 | 77 | ILRGSVAHK | Influenza A (PR8) NP 265-274 |
| Cancer | A*11:01 | 785 | VVGAVGVGK | KRAS G12V (8-16) |
| Cancer | A*11:01 | 3070 | VVVGADGVGK | KRAS G12D (7-16) |
| Cancer | A*11:01 | 3072 | VVVGAVGVGK | KRAS G12V (7-16) |
| COVID-19 / SARS-CoV-2 | A*11:01 | 4356B | KTFPPTEPK | SARS-CoV-2 Nucleocapsid protein 362-370 (confirmed epitope) |
| COVID-19 / SARS-CoV-2 | A*11:01 | 4679B | SVYAWNRKR | SARS-CoV-2 Spike glycoprotein 349-357 |
| COVID-19 / SARS-CoV-2 | A*11:01 | 4691B | VTNNTFTLK | SARS-CoV-2 ORF1ab 808-816 |
| Dengue | A*11:01 | 4632 | ATVLMGLGK | DENV 2 Polyprotein 2315-2323 |
| Dengue | A*11:01 | 3459 | ATYGWNLVK | DENV 1 NS5 2610-2618 |
| Dengue | A*11:01 | 3907 | GTSGSPIADK | Dengue NS3 133-142 |
| Dengue | A*11:01 | 2372 | GTSGSPIIDK | Dengue NS3 133-142 |
| Dengue | A*11:01 | 4626 | GTSGSPIINK | DENV 4 NS3 1607-1617 |
| Dengue | A*11:01 | 1843 | GTSGSPIINR | Dengue NS3 serotype 3&4 133-142 |
| Dengue | A*11:01 | 4625 | GTSGSPIVDK | DENV 2 NS3 1608-1617 |
| Dengue | A*11:01 | 3906 | GTSGSPIVDR | Dengue NS3 serotype 2 133-142 |
| Dengue | A*11:01 | 2373 | GTSGSPIVNR | NS3 serotype 1 133-142 |
| Dengue | A*11:01 | 3929B | KITAEWLWK | Dengue NS5 375-383 |
| Dengue | A*11:01 | 4630 | KTFDSEYIK | DENV 2 NS3 1863-1871 |
| Dengue | A*11:01 | 4629 | KTFDSEYVK | DENV 2 Polyprotein 1863-1871 |
| Dengue | A*11:01 | 4631 | KTFDTEYQK | DENV 1 NS3 1864-1872 |
| Dengue | A*11:01 | 3916B | RIEPSWADVK | Dengue NS3 64-74 |
| Dengue | A*11:01 | 3921B | RVIDPRRCMK | Dengue NS3 422-431 |
| Dengue | A*11:01 | 3910B | RVSTVQQLTK | Dengue C 22-31 |
| Dengue | A*11:01 | 3460 | STYGWNIVK | DENV 3 NS5 2607-2615 |
| Dengue | A*11:01 | 3461 | STYGWNLVR | DENV 2 NS5 2608-2616 |
| Dengue | A*11:01 | 4627 | TIPPTAGILK | DENV 2 Core 58-67 |
| Dengue | A*11:01 | 4628 | TTLSRTSKK | DENV 2 NS2a 1336-1344 |
| EBV | A*11:01 | 639 | ATIGTAMYK | EBV BRLF1 134-142 |
| EBV | A*11:01 | 726 | AVFDRKSDAK | EBNA3B 399-408 |
| EBV | A*11:01 | 199 | IVTDFSVIK | EBV EBNA-4 416-424 |
| EBV | A*11:01 | 508 | SSCSSCPLSK | EBV LMP-2 340-349 |
| EBV | A*11:01 | 4183 | SSCSSCPLTK | EBV LMP-2_EBVG 339-348 |
| HBV | A*11:01 | 3941 | STLPETTVVRR | Core 141-151 |
| HBV | A*11:01 | 2659 | YVNTNMGLK | Core 88-96 |
| HBV | A*11:01 | 377 | YVNVNMGLK | HBV core antigen 88-96 |
| HIV/SIV | A*11:01 | 1686 | ACQGVGGPGHK | HIV gag p24 |
| HIV/SIV | A*11:01 | 1486 | AVDLSHFLK | HIV nef 84-92 |
| HPV | A*11:01 | 2067 | NTLEQTVKK | HPV 33E6 86-94 |
| Influenza | A*11:01 | 1722 | KSMREEYRK | Influenza A MP2 70-78 |
| Influenza | A*11:01 | 1720 | RMVLASTTAK | Influenza A MP1 178-187 |
| Influenza | A*11:01 | 1513 | SIIPSGPLK | Influenza A MP 13-21 |
| Adenovirus | A*24:02 | 684 | TYFSLNNKF | Adenovirus 5 Hexon 37-45 |
| Cancer | A*24:02 | 230 | AFLPWHRLF | Tyrosinase 188-196 |
| Cancer | A*24:02 | 2302 | AYACNTSTL | Survivin 80-88 |
| Cancer | A*24:02 | 2303 | CYASGWGSI | Prostate Specific Antigen-1 153-161 |
| Cancer | A*24:02 | 3351 | DYLNEWGSRF | CDH3 807-816 |
| Cancer | A*24:02 | 1064 | DYLQYVLQI | MiHA ACC1 15-23 |
| Cancer | A*24:02 | 3353 | EYCPGGNLF | MELK 87-95 (93N) |
| Cancer | A*24:02 | 416 | EYLQLVFGI | MAGEA2 156-164 |
| Cancer | A*24:02 | 3380 | EYRALQLHL | CA9 219-227 |
| Cancer | A*24:02 | 3354 | EYYELFVNI | DEP DC1 294-302 |
| Cancer | A*24:02 | 3360 | GYCTQIGIF | HENMT1 221-229 |
| Cancer | A*24:02 | 237 | IMPKAGLLI | MAGE-A3 |
| Cancer | A*24:02 | 3362 | IYTWIEDHF | FOXM1 262-270 |
| Cancer | A*24:02 | 3367 | NYQPVWLCL | RNF43 721-729 (722Y) |
| Cancer | A*24:02 | 1806 | RYCNLEGPPI | Lymphocyte antigen 6 complex locus K (LY6K) 177-186 |
| Cancer | A*24:02 | 3372 | RYNAQCQETI | Midkine 110-119 |
| Cancer | A*24:02 | 3382 | SYRNEIAYL | TTK protein kinase 551-559 |
| Cancer | A*24:02 | 421 | TFPDLESEF | MAGEA3 97-105 |
| Cancer | A*24:02 | 209 | TYACFVSNL | Carcinogenic Embryonic Antigen (CEA) 652-660 |
| Cancer | A*24:02 | 417 | TYLPTNASL | HER-2/neu 63-71 |
| Cancer | A*24:02 | 238 | VYFFLPDHL | gp100-intron 4 (170-178) |
| Cancer | A*24:02 | 418 | VYGFVRACL | Telomerase reverse transcriptase (hTRT) 461-469 |
| Cancer | A*24:02 | 3384 | VYLRVRPLL | KIF20A 67-75 |
| Cancer | A*24:02 | 3385 | VYYNWQYLL | IL13r 146-154 |
| CMV | A*24:02 | 3390 | AYAQKIFKI | CMV IE-1 248-256 |
| CMV | A*24:02 | 414 | QYDPVAALF | HCMV pp65 341-349 |
| CMV | A*24:02 | 380 | VYALPLKML | HCMV pp65 113-121 |
| COVID-19 / SARS-CoV-2 | A*24:02 | 4692 | NYNYLYRLF | SARS-CoV-2 Spike glycoprotein 448-456 |
| COVID-19 / SARS-CoV-2 | A*24:02 | 4668 | QYIKWPWYI | SARS-CoV-2 Spike glycoprotein 1205-1213 |
| COVID-19 / SARS-CoV-2 | A*24:02 | 4670 | VYIGDPAQL | SARS-CoV-2 ORF1ab 5724-5732 |
| Dengue | A*24:02 | 2954 | QYSDRRWCF | Dengue NS3 557-565 (Singapore/S275/1990) |
| Dengue | A*24:02 | 4633 | RYLEFEALGF | DENV 3 NS5 2971-2980 |
| Dengue | A*24:02 | 4634 | TYFQRVLIF | DENV 2 M 261-269 |
| Dengue | A*24:02 | 4081 | TYGWNIVKL | DENV 3 NS5 2608-2616 |
| Dengue | A*24:02 | 4083 | TYGWNLVKL | DENV 1, 4 NS5 2611-2619 |
| Dengue | A*24:02 | 4082 | TYGWNLVRL | DENV 2 NS5 2609-2617 |
| EBV | A*24:02 | 648 | PYLFWLAAI | EBV LMP2 131-139 |
| EBV | A*24:02 | 420 | TYGPVFMCL | EBV LMP-2 419-427 |
| EBV | A*24:02 | 683 | TYGPVFMSL | EBV LMP2 419-427 |
| HBV | A*24:02 | 378 | EYLVSFGVW | HBV core 117-125 |
| HBV | A*24:02 | 3935 | FFPSIRDLL | core protein 23-31 |
| HBV | A*24:02 | 413 | KYTSFPWLL | HBV polymerase 756-764 |
| HCV | A*24:02 | 415 | AYSQQTRGL | HCV NS3 1031-1039 |
| HIV/SIV | A*24:02 | 419 | RYLKDQQLL | HIV-1 gag gp41 67-75 |
| HIV/SIV | A*24:02 | 1805 | RYLRDQQLL | HIV env |
| HIV/SIV | A*24:02 | 3238 | RYPLTFGW | HIV nef |
| HIV/SIV | A*24:02 | 3099 | RYPLTFGWCF | HIV NEF 143-152 |
| HIV/SIV | A*24:02 | 411 | RYPLTFGWCY | HIV-1 Nef 134-143 |
| HPV | A*24:02 | 3240 | VYDFAFRDL | HPV16 E6 |
| HTLV | A*24:02 | 1043 | SFHSLHLLF | HTLV Tax 301-309 |
| Cancer | A*29:02 | 3397 | KEKYIDQEEL | HSP90 alpha 280-288 (Pathologic Conditions) |
| HIV/SIV | A*29:02 | 3392 | LYNTVATLY | HIV gag 79-86 |
| HIV/SIV | A*29:02 | 3395 | SFDPIPIHY | HIV env 216-224 |
| HIV/SIV | A*29:02 | 3396 | SFNCRGEFFY | HIV env 382-391 |
| AAV | B*07:02 | 651 | VPQYGYLTL | AAV2 372-380 |
| Adenovirus | B*07:02 | 989 | KPYSGTAYNAL | Adenovirus Hexon 114-124 |
| Adenovirus | B*07:02 | 685 | KPYSGTAYNSL | AdV Hexon |
| BK virus | B*07:02 | 2070 | LPLMRKAYL | LT antigen 27-35 |
| Cancer | B*07:02 | 3348 | APRGVRMAV | LAGE-1 46-54 |
| Cancer | B*07:02 | 2369 | EPR(PHOSPHO-S)PSHSM | Insulin receptor substrate 2 |
| Cancer | B*07:02 | 3364 | LPVSPRLQL | CEACAM 185-193 |
| Cancer | B*07:02 | 2304 | LPWHRLFLL | Tyrosinase 208-216 |
| Cancer | B*07:02 | 3378 | SPFFLLLLL | Tumor Mucin antigen 7-15 |
| Cancer | B*07:02 | 3239 | TPNQRQNVC | P2X5 |
| CMV | B*07:02 | 308 | RPHERNGFTVL | HCMV pp65 265-275 |
| CMV | B*07:02 | 45 | TPRVTGGGAM | HCMV pp65 417-426 |
| COVID-19 / SARS-CoV-2 | B*07:02 | 4694 | RARSVSPKL | SARS-CoV-2 ORF7a 78-86 |
| COVID-19 / SARS-CoV-2 | B*07:02 | 4351 | SPRWYFYYL | SARS-CoV-2 Nucleocapsid protein 105-113 (confirmed epitope) |
| Dengue | B*07:02 | 3909 | APTRVVAAEM | Dengue NS3 serotype 2 222-231 |
| EBV | B*07:02 | 2305 | QPRAPIRPI | EBV EBNA-3C 881-889 |
| EBV | B*07:02 | 44 | RPPIFIRRL | EBV EBNA-3A 247-255 |
| EBV | B*07:02 | 647 | RPQGGSRPEFVKL | EBV BMRF1 116-128 |
| HAV | B*07:02 | 2164 | HPRLAQRIL | Hepatitis A Virus polyprotein 1877-1885 |
| HBV | B*07:02 | 701 | LPSDFFPSV | Core 19-27 |
| HCV | B*07:02 | 845 | DPRRRSRNL | HCV core 111-119 |
| HCV | B*07:02 | 742 | GPRLGVRAT | HCV core 41-49 |
| HIV/SIV | B*07:02 | 877 | GPGHKARVL | HIV gag p24 223-231 |
| HIV/SIV | B*07:02 | 196 | IPRRIRQGL | HIV-1 env gp120 848-856 |
| HIV/SIV | B*07:02 | 257 | TPGPGVRYPL | HIV-1 nef 128-137 |
| HPV | B*07:02 | 2065 | KPTLKEYVL | HPV 33 E7 5-13 |
| HTLV | B*07:02 | 3400 | LPVSCPEDL | bZIP factor 10-18 |
| Influenza | B*07:02 | 1743 | QPEWFRNVL | Influenza A PB1 329-337 |
| Influenza | B*07:02 | 1744 | SPIVPSFDM | Influenza A NP 473-481 |
| Minor Histocompatibility Antigen | B*07:02 | 1113 | SPSVDKARAEL | MiHAg SMCY 1041-1051 |
| Cancer | B*08:01 | 94 | GFKQSSKAL | bcr-abl 210 kD fusion protein 19-27 |
| CMV | B*08:01 | 3391 | ELKRKMIYM | CMV IE-1 199-207 (R/L Position 3, M/I Position 7) |
| CMV | B*08:01 | 127 | ELRRKMMYM | IE1 199-207 |
| Dengue | B*08:01 | 3925 | LEKTKKDL | Dengue NS4a 6-13 |
| Dengue | B*08:01 | 3915 | RIKQKGIL | Dengue NS3 25-32 |
| EBV | B*08:01 | 73 | FLRGRAYGL | EBV EBNA-3A 193-201 |
| EBV | B*08:01 | 489 | QAKWRLQTL | EBV EBNA3A 158-166 |
| EBV | B*08:01 | 190 | RAKFKQLL | EBV BZLF-1 190-197 |
| HBV | B*08:01 | 3937 | GLKILQLL | external core Ag |
| HCV | B*08:01 | 721 | HSKKKCDEL | HCV NS3 1395-1403 |
| HIV/SIV | B*08:01 | 879 | EIYKRWII | HIV p24 gag 128-135 |
| HIV/SIV | B*08:01 | 193 | FLKEKGGL | HIV-1 nef 90-97 |
| HIV/SIV | B*08:01 | 195 | GEIYKRWII | HIV-1 gag p24 261-269 |
| HIV/SIV | B*08:01 | 1536 | YLKDQQLL | Env 586-593 |
| Dengue | B*15:01 | 3912 | CLIPTAMAF | Dengue C 107-115 |
| HIV/SIV | B*15:01 | 1072 | RLRPGGKKKY | HIV-1 p17 20-29 |
| Cancer | B*27:05 | 424 | GRFGLATEK | BRAF 594-601 (600E) |
| Cancer | B*27:05 | 3359 | GRFGLATVK | BRAF 594-601 (600V) |
| Cancer | B*27:05 | 202B | RMFPNAPYL | WT-1 126-134 (Wilms tumor) |
| Chlamydia | B*27:05 | 3094 | ARKLLLDNL | PqqC-like protein 70-78 |
| Chlamydia | B*27:05 | 3945 | NRAKQVIKL | Probable ATP-dependent Clp protease ATP-binding subunit 7-15 |
| COVID-19 / SARS-CoV-2 | B*27:05 | 4354 | QRNAPRITF | SARS-CoV-2 Nucleocapsid protein 9-17 |
| HCV | B*27:05 | 871 | ARMILMTHF | NS5B 2841-2849 |
| HIV/SIV | B*27:05 | 2306 | GRAFVTIGK | HIV-1 gp100 103-111 |
| HIV/SIV | B*27:05 | 294 | KRWIILGLNK | HIV-1 gag p24 265-274 |
| HIV/SIV | B*27:05 | 874 | KRWIIMGLNK | HIV-1 Gag p24 263-272 |
| Influenza | B*27:05 | 463 | SRYWAIRTR | Influenza A NP 383-391 |
| Adenovirus | B*35:01 | 1346 | IPYLDGTFY | AdV Hexon |
| Cancer | B*35:01 | 2307 | MPFATPMEA | NY-ESO-1 94-102 |
| CMV | B*35:01 | 114 | IPSINVHHY | HCMV pp65 123-131 |
| Dengue | B*35:01 | 3908 | TPEGIIPTL | Dengue NS3 500-508 |
| EBV | B*35:01 | 399 | EPLPQGQLTAY | EBV BZLF-1 54-64 |
| EBV | B*35:01 | 3033 | HPVAEADYFEY | EBV EBNA-1 407-417(4A) |
| EBV | B*35:01 | 107 | YPLHEQHGM | EBV EBNA-3A 458-466 |
| HCV | B*35:01 | 942 | CPNSSIVY | HCV E1 207-214 |
| HCV | B*35:01 | 565 | HPNIEEVAL | HCV NS3 1359-1367 |
| HIV/SIV | B*35:01 | 255 | NPDIVIYQY | HIV-1 HIV-1 RT 328-336 |
| HIV/SIV | B*35:01 | 1211 | VPLDEDFRKY | HIV-1 HIV-1 RT 273-282 |
| COVID-19 / SARS-CoV-2 | B*40:01 | 4328 | MEVTPSGTWL | SARS-CoV-2 Nucleocapsid protein 322-330 (confirmed epitope) |
| Dengue | B*40:01 | 3924 | GEARKTFVEL | Dengue NS3 528-537 |
| EBV | B*40:01 | 692 | IEDPPFNSL | EBV LMP2 200-208 |
| HCV | B*40:01 | 939 | REISVPAEIL | HCV NS5a 2266-2275 |
| HIV/SIV | B*40:01 | 840 | KEKGGLEGL | HIV-1 Nef 92-100 |
| Other | E*01:01 | 712 | VMAPRTLIL | HLA-C leader sequence peptide |
| Other | E*01:01 | 2355 | VMAPRTLVL | HLA-A leader sequence peptide |
| Adenovirus | H-2Db | 2206 | SGPSNTPPEI | Ad5 E1A 234-243 |
| Autoimmunity | H-2Db | 1107 | RSPFSRVVHL | MOG Precursor 69-78 |
| Cancer | H-2Db | 695 | Abu-Abu-L-Abu-LTVFL | Moloney murine sarcoma virus (MoMSV) GagL 85-93 |
| Cancer | H-2Db | 2755 | AQLANDVVL | MC-38 |
| Cancer | H-2Db | 3349 | ASFRNLTHL | TPBG 258-266 |
| Cancer | H-2Db | 2695 | ASMTNMELM | MC-38 Adpgk neoantigen |
| Cancer | H-2Db | 1731 | ATFKNWPFL | Murine Survivin 20-28 |
| Cancer | H-2Db | 231 | CMTWNQMNL | WT1 235-243 |
| Cancer | H-2Db | 2199 | FSNSTNDILI | VEGFR2/KDR fragment 1 614-624 |
| Cancer | H-2Db | 559 | HCIRNKSVI | PSA 65-73 |
| Cancer | H-2Db | 1780 | HCIRNKSVIL | hPSA |
| Cancer | H-2Db | 1333 | KVPRNQDWL | gp100 (pmel17) 25-33 |
| Cancer | H-2Db | 202C | RMFPNAPYL | WT-1 126-134 (Wilms tumor) |
| Cancer | H-2Db | 2675 | STHVNHLHC | Endophilin-B2 244-252 |
| Cancer | H-2Db | 2198 | VILTNPISM | VEGFR2 400-408 |
| Cancer | H-2Db | 2415 | YSSDNLYQM | Wilms Tumor protein (221- 229) |
| Chlamydia | H-2Db | 3943 | ASFVNPIYL | Cysteine-rich membrane protein 63-71 |
| Chlamydia | H-2Db | 3947 | SAVSNLFYV | POMP 612-620 |
| CMV | H-2Db | 2313 | HGIRNASFI | MCMV m45 985-993 |
| HCV | H-2Db | 791 | GAVQNEVTL | HCV NS3 1629-1637 |
| HIV/SIV | H-2Db | 719 | AAVKNWMTQTL | SIV gag |
| HPV | H-2Db | 3404 | AGVDNRECI | HPV L1 165-173 |
| HPV | H-2Db | 502H | RAHYNIVTF | HPV 16 E7 49-57 |
| HSV | H-2Db | 3405A | AGPHNDMEI | HSV p56 487-495 |
| Influenza | H-2Db | 9 | ASNENMDAM | Influenza A (NT60) NP 366-374 |
| Influenza | H-2Db | 119 | ASNENMETM | Influenza A (PR8) NP 366-374 |
| Influenza | H-2Db | 3410 | SCLENFRAYV | Influenza A polymerease PA 224-233 |
| Influenza | H-2Db | 120 | SSLENFRAYV | Influenza A (PR8) Polymerase acidic protein 224-233 |
| LCMV | H-2Db | 70 | FQPQNGQFI | LCMV NP 396-404 |
| LCMV | H-2Db | 69 | KAVYNFATC | LCMV GP1 33-41 |
| LCMV | H-2Db | 142 | KAVYNFATM | gp33 (C9M) |
| LCMV | H-2Db | 186 | SGVENPGGYCL | LCMV GP 276-286 |
| Malaria | H-2Db | 2697 | SQLLNAKYL | Plasmodium berghei ANKA acid phosphatase 40-48 |
| Minor Histocompatibility Antigen | H-2Db | 328 | KCSRNRQYL | miHAg SMCY 738-746 |
| Minor Histocompatibility Antigen | H-2Db | 327 | WMHHNMDLI | miHAg UTY 246-254 |
| Tuberculosis | H-2Db | 547 | AIQGNVTSI | Mycobacterium tuberculosis ESAT-6 17-25 |
| West Nile | H-2Db | 663 | LGMSNRDFL | West Nile Virus polyprotein 294-302 |
| CMV | H-2Dd | 278 | AGPPRYSRI | MCMV m164 257-265 |
| CMV | H-2Dd | 2167 | LGPISGHVL | pp65 15-23 |
| HIV/SIV | H-2Dd | 1684 | IGPGRAFYA | HIV-1 US4 gp120 318-326 |
| HIV/SIV | H-2Dd | 177 | RGPGRAFVTI | HIV-1 IIIB gp120 318-327 |
| HSV | H-2Dd | 3405B | AGPHNDMEI | HSV p56 487-495 |
| Cancer | H-2Kb | 351 | HNTQYCNL | MAGEA5 5-12 |
| Cancer | H-2Kb | 3373 | SDYYFSWL | muFAP 289-296 |
| Cancer | H-2Kb | 2754 | SIIVFNLL | MC-38 |
| Cancer | H-2Kb | 3376 | SKYVFENV | Mybpc2 295-302 |
| Cancer | H-2Kb | 185 | SVYDFFVWL | Tyrosinase related protein-2 180-188 |
| Cancer | H-2Kb | 2311 | TVSEFLKL | Murine Survivin 97-104 |
| Cancer | H-2Kb | 2585 | VGRNFTNL | mTERT |
| CMV | H-2Kb | 1915 | TVYGFCLL | MCMV M139 419-426 |
| COVID-19 / SARS-CoV-2 | H-2Kb | 4549 | SIIAYTMSL | SARS-CoV-2 Spike glycoprotein 691-699 |
| COVID-19 / SARS-CoV-2 | H-2Kb | 4544 | VVLSFELL | SARS-CoV-2 Spike glycoprotein 505-523 |
| HBV | H-2Kb | 422 | ILSPFLPLL | HBV surface antigen 208-216 |
| HBV | H-2Kb | 1431 | MGLKFRQL | HBV core 93-100 |
| HBV | H-2Kb | 477 | VWLSVIWM | HBV Surface antigen 190-197 |
| HPV | H-2Kb | 2560 | EVYDFAFRDL | HPV E6 48-57 |
| HSV | H-2Kb | 188 | SSIEFARL | HSV-1 gp B 498-505 |
| LCMV | H-2Kb | 2076 | ISHNFCNL | LCMV GP 118-125 |
| Model Antigens | H-2Kb | 908 | KVVRFDKL | Ovalbumin (subdominant) 55-62 |
| Model Antigens | H-2Kb | 562 | LTFNYRNL | Histocompatibility antigen 60 39-46 |
| Model Antigens | H-2Kb | 93 | SIINFEKL | Ovalbumin 257-264 |
| Model Antigens | H-2Kb | 1803 | SIYRYYGL | Synthetic variant of VSV Nucleoprotein 498-505 |
| MuLV | H-2Kb | 828 | KSPWFTTL | MuLV env 622-629 (MC38) |
| Other | H-2Kb | 2331 | TSINFVKI | MHV 524-431 |
| Sendai Virus | H-2Kb | 56 | FAPGNYPAL | Sendai virus NP 324-332 |
| SV40 | H-2Kb | 488 | VVYDFLKL | SV40 T antigen 404-411 |
| Toxoplasma gondii | H-2Kb | 2156 | SVLAFRRL | tgd057 57-64 |
| Trypanosoma Cruzi | H-2Kb | 708 | ANYNFTLV | Trypanosoma cruzi SP 536-543 |
| Trypanosoma Cruzi | H-2Kb | 964 | VNHRFTLV | Trypanosoma cruzi ASP-2 553-560 |
| Tuberculosis | H-2Kb | 764 | IMYNYPAM | Mycobacterium tuberculosis TB10.4 4-11 |
| Tuberculosis | H-2Kb | 769 | IMYNYPAML | Mycobacterium tuberculosis TB10.4 4-12 |
| Vaccinia | H-2Kb | 733 | TSYKFESV | Vaccinia virus WR epitope B8R 20-27 |
| Vesicular Stomatitis Virus | H-2Kb | 373 | RGYVYQGL | VSV NP 52 |
| Yellow Fever Virus | H-2Kb | 1182 | ATLTYRML | Yellow Fever Virus 17D polyprotein 268-275 |
| Cancer | H-2Kd | 485 | AYIDFEMKI | SART3 315-323 |
| Cancer | H-2Kd | 814 | EYILSLEEL | GPC3 298-306 |
| Cancer | H-2Kd | 3381 | SYMLQALCI | TNPO3 |
| Cancer | H-2Kd | 343 | TYVPANASL | Neu/Her-2/Erbb2 proto-oncoprotein 66-74 |
| CMV | H-2Kd | 3412 | CYYASRTKL | MCMV m145 451-459 |
| HBV | H-2Kd | 526 | SYVNTNMGL | Core 87-95 |
| HBV | H-2Kd | 529 | WYWGPSLYSI | large envelope 362-371 |
| HIV/SIV | H-2Kd | 403 | AMQMLKDTI | HIV gag p24 197-205 |
| HIV/SIV | H-2Kd | 176 | AMQMLKETI | HIV-1 gag p24 199-207 |
| HSV | H-2Kd | 3407 | TYWPVVSDI | 207-215 |
| Influenza | H-2Kd | 240 | IYSTVASSL | Influenza A HA 533-541 |
| Influenza | H-2Kd | 98 | TYQRTRALV | Influenza A (PR8) NP 147-155 |
| Listeria | H-2Kd | 178 | GYKDGNEYI | Listeria monocytogenes Listeriolysin 91-99 |
| Malaria | H-2Kd | 376 | SYIPSAEKI | Plasmodium berghei CSP 252-260 |
| Malaria | H-2Kd | 2290 | SYVPSAEQI | Plasmodium CSP |
| Malaria | H-2Kd | 771 | YYIPHQSSL | Plasmodium falciparum Liver stage antigen 1671-1679 |
| Model Antigens | H-2Kd | 198 | HYLSTQSAL | Enhanced Green Fluorescent Protein (eGFP) 200-208 |
| Other | H-2Kd | 3414 | IYNVGQVSI | Leptospira |
| Other | H-2Kd | 458 | KYNKANVFL | NRP-V7 superagonist peptide 8.3 Tg NOD mouse |
| Other | H-2Kd | 62 | RYLKNGKETL | HLA-Cw3 170-179 |
| RSV | H-2Kd | 149 | KYKNAVTEL | RSV A strain F protein 85-93 |
| RSV | H-2Kd | 154 | SYIGSINNI | Respiratory Syncytial Virus (RSV) M2 82-90 |
| Tuberculosis | H-2Kd | 1263 | VYAGAMSGL | Mtb85A 145-152 |
| Autoimmunity | H-2Kd | 580 | VYLKTNVFL | IGRP (206-214) |
| Cancer | H-2Ld | 150 | LPYLGWLVF | Tumour Antigen P815 35-43 |
| CMV | H-2Ld | 21 | YPHFMPTNL | MCMV IE1 168-176 |
| HBV | H-2Ld | 129 | IPQSLDSWWTSL | HBV surface antigen 28-39 |
| Model Antigens | H-2Ld | 74 | TPHPARIGL | E. coli beta-galactosidase 876-884 |
| MuLV | H-2Ld | 398 | SPSYVYHQF | MuLV env gp70 423-431 |
| Tuberculosis | H-2Ld | 158 | MPVGGQSSF | Mycobacterium tuberculosis Ag85A 70-78 |
| Vesicular Stomatitis Virus | H-2Ld | 622 | MPYLIDFGL | VSV N 275-283 |
| Other | H-2Ld | 3051 | LPNWGKYVL | Rabies Glycoprotein (453-461) |
Custom ProM1™ Class I MHC Monomers – at no extra charge
Custom ProM1™ Monomers for your allele/peptide of interest are available within approximately 6-7 weeks from ordering. Peptide sequences are subject to prior evaluation by ProImmune for binding. Once a peptide sequence is accepted for custom synthesis we will synthesize both the peptide and MHC monomer reagent. Custom ProM1™ Monomers are provided at no extra charge compared to catalog ProM1™ Monomers.
Available Alleles
HLA-A*01:01, A*02:01, A*03:01, A*11:01, A*24:02
HLA-B*07:02, B*08:01, B*35:01
HLA-E*01:01
H-2 Kb, Kd, Db, Ld
If your allele is not on the list above, please enquire.
