Your basket is currently empty!
ProImmune REVEAL & ProVE® Overview
ProImmune REVEAL® MHC-Peptide Binding Assays
find out what you need to know with the world’s most cited MHC-peptide binding assay service
In vitro MHC peptide-binding assays do what prediction tools can’t. They confirm the physical binding characteristics of actual peptides, based on their sequence.
ProImmune is the world leader in providing in vitro MHC peptide binding assays, offering the widest range of MHC alleles and assay options, from high throughput screening to in-depth competition assays between candidate peptides. Our specialist immunology advisory team can help you define your project needs and quickly tailor the most relevant assay solution.
ProImmune REVEAL® data published by researchers at US FDA:
Researchers at the US FDA and colleagues use ProImmune REVEAL to understand how immunogenicity of therapeutic Factor VIII protein varies between ethnic groups based on their HLA tissue-type (Pandey G.S. et al. PLOS Computational Biology May 2013, Vol. 9 (5) e1003066):
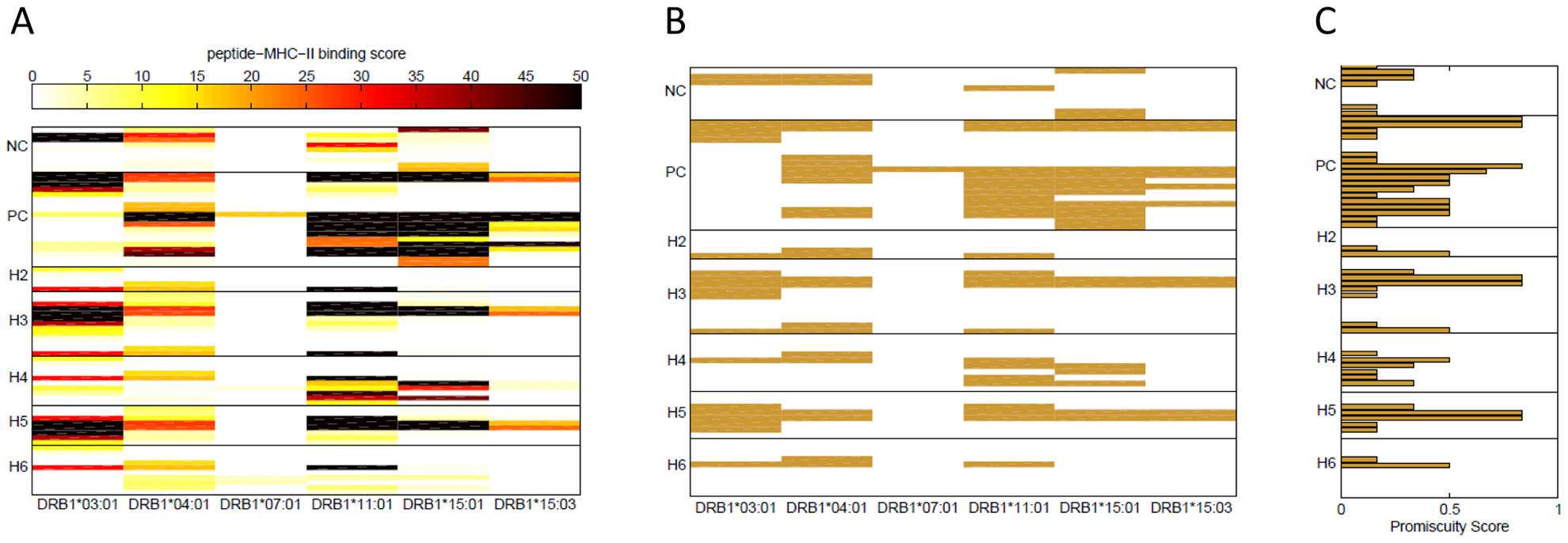
Figure. (A) Binding scores of FVIII wild type peptides. Binding scores in a REVEAL assay were measured for the synthetic wild type peptides depicted in bold in Table 1. Each row represents a peptide and each column a MHC-II allele. The peptides are grouped together based on the position in the FVIII gene where the haplotypes H2 to H6 occur. Overlapping peptides were used around the position of each ns-SNP. Positive-control (PC) peptides are known T-cell epitopes of FVIII and negative control (NC) peptides are from regions of the FVIII protein where no T-cell epitopes have been identified. The scale for the binding score is shown at the top of the figure; dark colors represent peptides that bind with high affinity to the MHC-II allele. (B) FVIII high affinity binders. Wild type peptides with binding score >15% are shown and considered potential T-cell epitopes for each of the six MHC-II alleles. (C) Binding promiscuity scores for the wild type FVIII peptides. The binding promiscuity score for each peptide, defined as the fraction of MHC-II variants the peptide binds with a score >15%, is shown.
ProImmune REVEAL® is widely applicable across many disease areas including all areas of cancer and infectious diseases. New CD4+ and CD8+ T cell epitopes identified with the help of this technology can be used to assess protein antigenicity or as core building blocks for vaccine development or as targets for new immunotherapy.
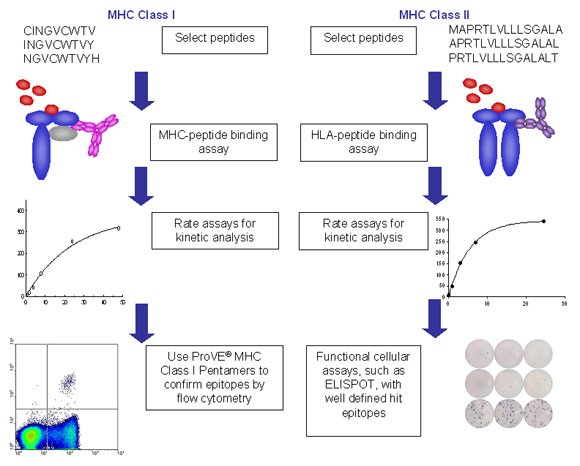
Figure 2. Example process flows for ProImmune REVEAL assays for class I and class II MHC.
- In each case peptides are synthesized for the relevant project, based on ProImmune’s versatile thinkpeptides peptide synthesis platform. Peptides used can be in any relevant form, whether from overlapping sequences or individually selected peptides.
- Peptides are then incubated in each case with recombinant MHC molecules and the stabilization of the MHC peptide complex is measured in an immunoassay format.
- Assembly and dissociation can be measured in high throughput at a single time interval in each case, allowing many alleles and peptides to be screened in parallel and an affordable cost.
- Alternatively, or as a follow-up for peptides of particular interest, more in-depth rate assays can be performed to establish more detailed on and off rate characteristics.
- Epitope validation can be carried out either using MHC-multimers, such as ProVE® Pentamers for MHC class I or ProT2® Tetramers for MHC class II. As an alternative, or in combination with MHC multimer technology, functional assays, such as ELISpot, flow cytometry analysis of intracellular cytokine staining, or proliferation assays can be performed to obtain functional confirmation for specific peptides.
The full length presentation can be viewed here
Praise for ProImmune REVEAL®
Prof. Julio Delgado, University of Utah, USA “The advantage of ProImmune’s in vitro system is that we could characterize binding to each HLA molecule individually and not have to guess which of the range of HLA molecules expressed by our patients was responsible for antigen presentation”.
Dr. Gene Olinger, United States Army Medical Research Institute of Infectious Diseases (USAMRIID) “ProImmune accomplished in two months what would have taken three years in our laboratory”.
Key publication:
ProImmune REVEAL® used to identify novel CD20 specific CD4+ T cell epitopes relevant in lymphoma immunotherapy
Milcent B., Siberil S. et al, (2019), “Presence of T cells directed against CD20-derived peptides in healthy individuals and lymphoma patients” Scientific Reports (2019),Vol. 9, Art.No. 10165
Cancer Immunology, Immunotherapy
Abbreviated summary:
Preclinical and clinical studies have suggested that cancer treatment with antitumor antibodies induces a specific adaptive T cell response. A central role in this process has been attributed to CD4+ T cells. In this study human CD20-derived epitopes restricted by HLA-DR1, HLA-DR3, HLA-DR4, and HLA-DR7 were characterized.
The results from this study indicate that carefully selected CD20-derived MHC II-restricted peptides make it possible to induce CD20-specific CD4+ T cell responses in humanized HLA-DR-transgenic mice and in human PBMCs. These peptides could serve as a therapeutic tool in B cell malignancies to improve the antitumor activity of CD4+ T cells in the context of vaccination strategies by helping CD8+ T cell response and eventually through direct cytotoxic effector functions.
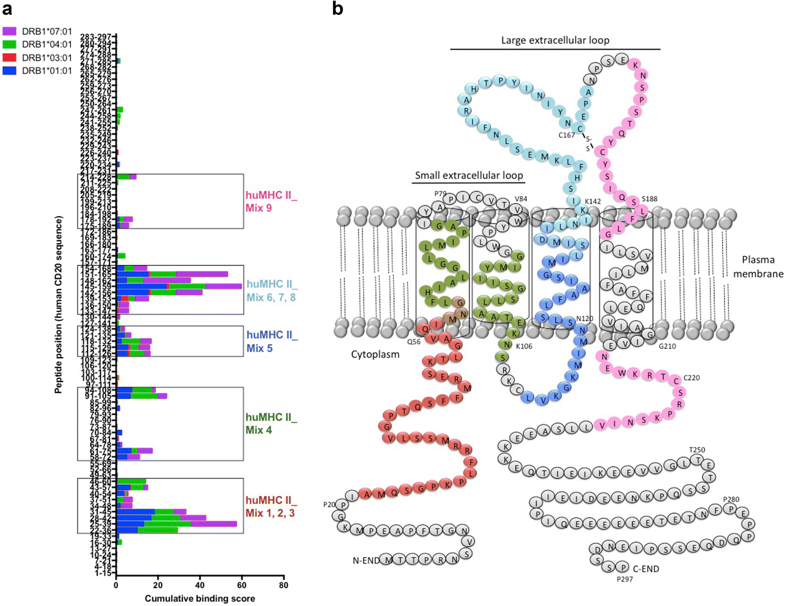
Figure. Screening of immunogenic HLA-DR-restricted CD20-derived peptides. a Cumulative scores of the binding of human CD20-derived peptides to recombinant HLA-DRB1*01:01 (blue), *03:01 (red), *04:01 (green), and *07:01 (purple) molecules as calculated with the ProImmune REVEAL® MHC-peptide binding assay. High scoring peptides within intracellular, transmembrane, and extracellular domains of the human CD20 molecule were pooled into 9 different mixtures of 18 to 20-mer MHC II-restricted peptides (huMHC II_Mix 1 to 9) (see also Supplementary Table 1). b Localization of the different MHC II-restricted CD20-derived peptide mixtures (huMHC II_Mix 1 to 3 in red; huMHC II_Mix 4 in green; huMHC II_Mix 5 in dark blue; huMHC II_Mix 6 to 8 in light blue; huMHC II_Mix 9 in pink).
What will you get?
ProImmune REVAL® data is reported in our detailed reporting templates for this assay range. Reports present data from the particular assays selected.
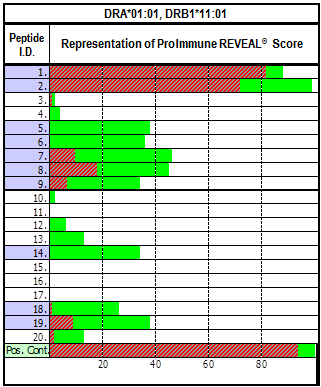 |
Figure. REVEAL Class II Example Data Presentation Binding as a percentage of the positive control is shown in green. Red overlay: Stability after 24hrs at 37°C. |
ProImmune REVEAL® Available MHC Alleles |
||
| Class I HLA
A*01:01 A*02:01 A*03:01 A*11:01 A*24:02 A*29:02 B*07:02 B*08:01 B*14:02 B*15:01 B*27:05 B*35:01 B*40:01 Class I: H-2 H-2Kb H-2Db H-2Ld H-2Kd H-2Dd Class I: Mamu Mamu-A*01 Mamu-A*02 Class II: IA H-2IAb H-2IAd |
Class II: HLA DR
A1*01:01/B1*01:01 A1*01:01/B1*15:01 A1*01:01/B1*03:01 A1*01:01/B1*04:01 A1*01:01/B1*11:01 A1*01:01/B1*13:01 A1*01:01/B1*07:01 A1*01:01/B1*01:02 A1*01:01/B1*04:02 A1*01:01 /B1*04:04 A1*01:01/B1*04:05 A1*01:01/B1*04:07 A1*01:01 /B1*04:08 A1*01:01/B1*08:04 A1*01:01/B1*09:01 A1*01:01/B1*10:01 A1*01:01/B1*11:02 A1*01:01/B1*11:03 A1*01:01/B1*11:04 A1*01:01/B1*15:02 A1*01:01/B1*15:03 A1*01:01/B1*16:01 A1*01:01/B1*16:02 A1*01:01/B3*02:02 A1*01:01/B3*03:01 A1*01:01/B5*01:01 |
Class II: HLA DQ
A1*01:01/B1*05:01 A1*05:01/B1*03:01 A1*01:02/B1*05:02 A1*01:02/B1*06:02 A1*03:01/B1*03:02 A1*01:02/B1*06:04 A1*05:01/B1*02:01 A1*02:01/B1*02:02 A1*03:01/B1*03:01 Class II: HLA DP A1*01:03/B1*01:01 A1*01:03/B1*02:01 A1*01:03/B1*03:01 A1*01:03/B1*04:01 A1*01:03/B1*04:02 A1*01:03/B1*05:01 A1*02:01/B1*01:01 A1*02:01/B1*02:01 A1*02:01/B1*03:01 A1*02:01/B1*04:01 A1*02:01/B1*04:02 A1*02:01/B1*05:01 A1*02:01/B1*06:01 A1*02:01/B1*09:01 A1*02:01/B1*11:01 A1*02:01/B1*13:01 A1*02:01/B1*14:01 A1*02:01/B1*15:01 A1*02:01/B1*17:01 A1*02:02/B1*05:01 |
High throughput ProImmune REVEAL® data for Remicade®
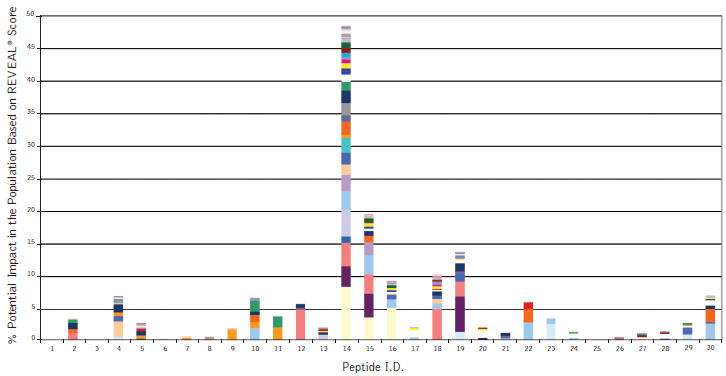
Figure: ProImmune REVEAL® results showing the population impact of Class II HLA binding peptides representing part of the mouse variable region of Remicade® (Infliximab). Remicade, a highly successful drug for the treatment of inflammatory autoimmune diseases also shows high rates of incidence of immunogenicity. The high-throughput nature of ProImmune REVEAL® allows researchers to develop a comprehensive understanding of how entire domains of monoclonal antibodies or therapeutic proteins interact with HLA molecules from a global population group based on actual experimental binding studies.
ProImmune REVEAL® Applications
- Rapid, comprehensive epitope mapping, resolving MHC binding restriction experimentally.
- Study of the immunogenicity of different variants of a protein, e.g. comparison of the variable and conserved regions of immunogenic proteins in different clades of a virus
- Optimization of clinical monitoring e.g. selection of the optimal Pentamer specificities for monitoring a target patient group
- Identification of cross presentation of a peptide by several alleles
- Improvement in vaccine design and efficacy.
- Mutagenesis studies: epitope binding optimization for therapeutic application, such as the insertion of anchor residues in peptide sequences with only partial or weak anchors
ProImmune REVEAL® Advantages
- Physical assay. Experimental results overcome hypothetical nature of MHC-peptide binding prediction.
- Speed. Get experimental data quickly.
- Flexibility. Many options exist for our ProImmune REVEAL® system both for high throughput analysis of many peptides across a wide range of alleles to in-depth analysis for a smaller number of peptides on specific MHC types. Our immunology specialist sales team can help configure the study best suited to your objectives.
- Affordability. The scalability and flexibility of ProImmune REVEAL® means that a project can be designed to suit your budget objective. We can crystalize the most important questions to be answered and design projects that can be staged to manage program risks.
Improved Intellectual Property Position
Identification of the individual epitopes of a therapeutic protein or other protein of interest allows separation of the intellectual property (IP) of that protein from the IP around the method of delivery or its therapeutic use. During vaccine development, defining IP for specific epitopes increases the options for future changes in product design. If the IP for the specific antigens in a protein of interest is not secured, competitive organizations may have the opportunity to develop an obstructive patent position.
FAQ
Q: What information is required to get a quote for a ProImmune REVEAL project?
A: Ideally we need to know the peptide sequences you are interested along with the MHC alleles you want us to screen. We can however provide outline quotes for you based on numbers of peptides and alleles only, which will be a good start for scoping your project.
Q: How long does this service take?
A: This will very much depend on the project scope, including the number and specification of the peptides used. Once we know your project objective we will always provide you with an estimated delivery time with our quotes.
Q: With all the available options, how do I decide which one is best for me?
A: Based on the understanding that we develop in discussing your requirement with you, we will be able to propose the most suitable project scope for you and discuss how this can fit your broader study objectives and other assay work.
Case Study:
ProImmune REVEAL® & ProVE® Rapid Epitope Discovery System used to identify novel T cell epitopes in HIV-1
Westrop, S.J. et al. (2009). “Novel approach to recognition of predicted HIV-1 Gag B*35:01-restricted CD8 T-cell epitopes by HLA-B*35:01+ patients: Confirmation by quantitative ELISpot analyses and characterisation using multimers.” J Immunol Methods. 341: 76-85. [PubMedID: 19056394]
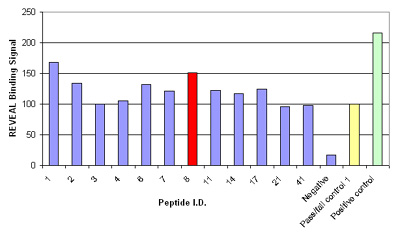
Figure. Results of the MHC-peptide binding assay for controls and the 12 peptides that were considered potential epitopes; binding signal is shown as a percentage relative to the pass/fail control; peptide 8, HPVHAGPIA (red) was further validated by Pentamer staining.
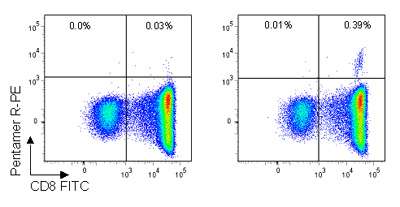
Figure. Pro5® Pentamer staining of live lymphocytes gated on CD3+ cells; the left plot shows staining with an allele mismatched negative control Pentamer, the right plot shows staining with the HA9 (B*35:01/HPVHAGPIA) Pentamer. 0.39% of CD3+ live lymphocytes are CD8+/HA Pentamer+ with a background stain of 0.03%.
The author commented that this study “confirms the novel ProImmune technology as a useful and superior tool for revealing potential immunogenic epitopes”, and also highlighted the importance of validating results in parallel using functional assays, such as ELISpot and Pro5® Pentamer staining.
Key Publication
See how Baxter used ProImmune REVEAL® Binding Assays to understand the immunogenicity of human Factor VIII treatment:
Steinitz, K. et al . “CD4+ T-cell epitopes associated with antibody responses after intravenously and subcutaneously applied human FVIII in humanized hemophilic E17 HLA-DRB1*15:01 mice.” Blood (2012) [PubMedID:22394599]