Your basket is currently empty!
Lessons-in-Developing
Lessons in Developing Effective Vaccine Strategies
Insights into Vaccine Discovery at Intercell AG

Dr. Sanja Selak (front row, second from left) and the Serology and Immune Assays team at Intercell AG, Austria.
Epitope discovery systems, such as ProImmune’s Antigen Characterization Platform, often identify a large number of candidate epitope sequences. While many protein sequences can potentially be antigenic only a few will be relevant as key antigens with addressable immune responses in vivo. To complicate matters further, the available antigens from pathogens may alter over the course of an infection, as Dr. Sanja Selak of Intercell AG, Austria, outlined in her presentation at ProImmune’s Antigen Characterization and Biomarker Discovery Summit in January 2011.
Dr. Selak is investigating the B cell epitopes of pathogenic bacteria, and their potential for use in vaccination strategies. Bacterial antigens are especially tricky to work with because bacteria do not express the same proteome consistently. They react to their environment, in particular the availability of micronutrients, and respond by expressing the cellular machinery they require to survive, so that a whole gamut of new antigens can suddenly become available. She has to consider not only the ‘who’ of candidate epitopes from her discovery strategies, but the ‘when’ ‘where’ and ‘why’, as bacteria adapt to survive in their local habitat.
The antigen identification strategy used at Intercell follows a multistep process, requiring many different assays and taking a sizable amount of time. Firstly, acute and convalescent serum pairs from patients who successfully recovered from infection are studied. High antibody titres suggest that the antigen was immunogenic, expressed during infection, and, most importantly, contributed to bacterial clearance. As comparators, the team use sera from healthy, disease-exposed individuals, and analyze the antibody repertoire of patients who fail to clear the infection.
Next, in vitro assays are brought in to play. The target antigen must be an accessible cell surface protein, and the antibodies from patient sera need to demonstrate high-affinity binding. The ‘antigenome’ is considered – are the candidates conserved between bacterial strains?
To validate their candidates, the research team at Intercell rely on knowledge of the immune evasion mechanisms specific to each pathogen to create custom assays. Human pathogens such as S. pyogenes, S. pneumoniae and P. aeruginosa survive in their hosts through expression of complement-neutralizing proteins, Fc binding proteins, extracellular matrix binding proteins, and host nutrient acquisition proteins. These adaptations to human hosts are not always apparent in straightforward in vitro cell culture experiments. In the example shown in figure 1, when P. aeruginosa is grown in normal cell culture conditions the D5 cell surface antigenic epitope is barely detectable, but adding 50% human serum causes up-regulation of D5 expression, validating it as a lead candidate.
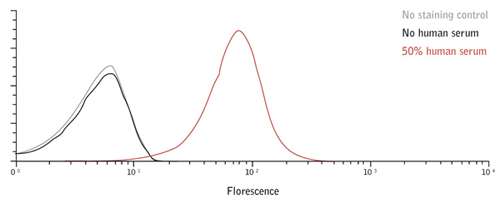
Figure 1: P. aeruginosa stained with an antibody to the DR5 antigen in the absence (black) and presence (red) of 50% human serum. When P. aeruginosa is grown in normal cell culture conditions the D5 cell surface antigenic epitope is barely detectable, but adding 50% human serum causes up-regulation of D5 expression, validating it as a lead candidate.
Other assay adaptations the team have explored include cell culture in iron-depleting conditions, and opsonization and cell killing assays performed with varied phagocytic cells in the presence of human or animal complement sources. Different combinations of phagocytic cells, complement sources and growth conditions are tested to identify the most optimal assay setup for testing bactericidal activity of antibodies against a particular pathogen; taking into account its immune evasion mechanisms.
Since animal models are of limited value with such well-adapted human pathogens, Dr. Selak and her colleagues are now beginning to consider the potential offered by tissue explant cultures for simulation of bacterial microenvironment during infection. Their knowledge–driven approaches to validating leads from each bacterium they investigate could yield effective vaccine strategies.
Intercell’s approach is enabling them to develop a new generation of vaccines that include only essential components and that stimulate both T cells and B cells. Complementary technologies, such as ProImmune’s Antigen Characterization Platform, can be used to enhance understanding of T and B cell responses and provide tools to track such responses at different stages of disease. In particular, the in vitro ProImmune REVEAL® MHC-peptide binding and rate assays, offer a rapid method for T cell epitope discovery, accelerating research projects and consequently saving time and money.
This work was carried out at Intercell AG, Austria
UPDATE: Intercell AG is Defunct as of 2013
