Your basket is currently empty!
MHC-Class-I-Pentamers
Case Study:
ProImmune Pro5® MHC Class I Pentamers are used to provide new insight into the selection of disease epitope-specific TCR
Gras, S. et al. (2010). Allelic polymorphism in the T cell receptor and its impact on immune responses. Journal of Experimental Medicine. 207(7): 1555-67. [PubMed ID 20566715]
Though many millions of T cell receptor (TCR) specificities are possible, there are often strong population biases in the presence of particular TCRs specific for disease epitopes. Using ProImmune Pro5® MHC Class I Pentamers, Gras et al. addressed the previously overlooked question of the impact of sequence variation in the TCR on immunity. They demonstrated that a single amino acid difference within a TCR can reduce its binding affinity for a viral peptide-MHC complex and preclude its dominance in an antiviral T cell response.
As their model system, the team chose to employ the HLA-B*35:01 – restricted HPVGEADYFEY (HPVG) epitope from the EBNA-1 protein of Epstein Barr Virus (EBV), which is highly immunogenic in EBV-exposed individuals. They established that the T cell response to this epitope is characterized by biases in TCR Va gene (TRAV and TRBV) use towards TRAV20 and TRBV9, suggesting that such recurrence is driven by specificity for the B*35:01/HPVG MHC-peptide complex. Genomic sequencing of the TRBV loci of 27 HLA-B*35:01+, EBV-exposed individuals identified 21 donors as homozygous for the TRBV9*01 allele, and 6 as heterozygous, also possessing TRBV9*02. Next, a Pro5® B*35:01/HPVGEADYFEY Pentamer was used to sort epitope-specific T cells, and their TRBV9 gene products were cloned for sequencing. All were found to be positive for TRBV9*01, indicating that only this allele is used in the response to HPVG.
To establish the structural basis for preferential TRBV9*01 selection, Jurkat human T cells were transduced to express TCR containing either TRBV9*01 wild type (WT), or one of two variants, Gln55His and Gln55Ala. The B*35:01/HPVG Pentamer was used to assess antigen recognition by these variants. At 4ºC, the WT TCR showed strong binding, while Gln55His stained poorly, indicating reduced avidity for MHC-peptide. Further, measurement of up-regulation of CD69 as a marker of activation after incubation with B*35:01/HPVG Pentamer showed that, unlike the WT and Gln55Ala variants, Gln55His cell lines were essentially unresponsive to cognate peptide-MHC: these data are shown in Figure 1.
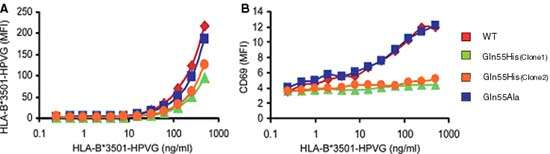 |
| Figure 1 A The TCR-transduced JurkatCD8 cells were also stained at 37°C for 4 h with the B*35:01-HPVG Pentamer using a range of multimer concentrations (final concentration indicated on the x axis). Mean fluorescence intensity (MFI) of HLA-B*3501/HPVG Pentamer staining is shown on the y axis. B Activation of the TCR-transduced JurkatCD8 cells with various concentrations of the HLA B*35:01/HPVG Pentamer. CD69 up-regulation was used as a marker for cell activation. The binding of HLA-B*35:01/HPVG Pentamer was performed at 37°C for 4 h. The cells were then washed and costained with APC-labeled anti-CD69 antibody for 30 min on ice. The experiments were conducted at least twice with similar results. |
Again using CD69 upregulation as a measure of activation, Gras et al. examined T cell recognition of HPVG epitope sequence variants from different strains of EBV. The WT and to a lesser extent the Gln55Ala TCRs were able to recognize both HPVGEADYFEY and HPVAEADYFEY, and make some response to HPVGQADYFEY, while the Gln55His mutation was far less responsive to all epitopes. Thus the WT TCR is finely tuned for specific recognition of the HPVG epitope, yet also recognizes viral variants. Using surface plasmon resonance, the team confirmed that Gln55His had a lower affinity for all HPVG variants than the WT.
Analysis of the structures of B*35:01/HPVGEADYFEY-WT, B*35:01/HPVGEADYFEY-Gln55Ala and B*35:01/HPVGEADYFEY-Gln55His TCR-pMHC crystals solved by Gras et al. consolidated their observations of the binding properties of these TCRs to B*35:01/HPVG. At the peptide-MHC/TCR interface in the region of polymorphic residue 55, the Gln55Ala substitution led to a loss of several key hydrogen bonds on the interaction surface. The Gln55His substitution leads to altered charge complementarity at the interaction surface: the presence of a bulky His residue, coupled with the loss of a polar Gln means that the Asp residue from HPVGEADYFEY is less readily accommodated by this TCR, so a weaker interaction and a faster off-rate ensue.
ProImmune Pro5® Pentamer technology thus proved a useful tool to this team of researchers, enabling them to correlate their structural data with observations of dominant TCR selection patterns. Fully understanding the TCR peptide-MHC interaction in the context of disease will be key to the successful development of novel therapeutics.
©Gras et al., 2010. Originally published in the Journal of Experimental Medicine doi:10.1084/jem20100603
