Your basket is currently empty!
ProImmune-REVEAL
Case Study:
ProImmune REVEAL® & ProVE® Rapid Epitope Discovery System used to identify novel T cell epitopes in HIV-1
Westrop, S.J. et al. (2009). Novel approach to recognition of predicted HIV-1 Gag B*35:01-restricted CD8 T-cell epitopes by HLA-B*35:01+ patients: Confirmation by quantitative ELISpot analyses and characterisation using multimers. J Immunol Methods. 341: 76-85. [PubMedID: 19056394]
The genetic profile of an individual, such as the patient’s tissue type, is understood to greatly influence the rate of HIV-1 disease progression. For example, HLA-B35 and -B8 have been associated with an increased rate of disease progression in HIV-1+ individuals. ProImmune’s REVEAL® & ProVE® Rapid Epitope Discovery System was used to investigate the HIV-1 Gag-specific CD8+ T cell immune response mounted by HLA-B35+ individuals to discover the possible reasons for this rapid advancement in the disease.
Westrop et al. used the MHC-peptide binding assay to screen 9-mer peptide sequences from the HIV-1 Gag protein against the HLA-B*35:01 allele. A previously unknown epitope was identified, which may be involved in an immune response in B*35:01-positive individuals.
ProImmune synthesized a PEPscreen® Custom Peptide Library of forty-four 9-mer HIV-1 Gag peptides. These peptides were assessed for binding to B*35:01 using the MHC-peptide binding assay. The binding affinity of the peptides was compared to a pass/fail control peptide known to have marginal affinity for the B*35:01 MHC complex. Peptides with a conformational signal >80% of the pass/fail control signal were considered to be potential epitopes (Figure 1). Twelve HIV-1 Gag peptides fulfilled this criterion, and these peptides were further analyzed using the Quick Check Off Rate Assay. The rates of dissociation of the peptide-MHC complexes were measured at 0, 2 and 24 hours and the half-lives of the complexes were calculated to give an indication of their overall stability. Two peptides, YPLTSLRSL (YL9) and HPVHAGPIA (HA9) were found to form a stable complex with B*35:01, indicating that these may be potential novel epitopes.
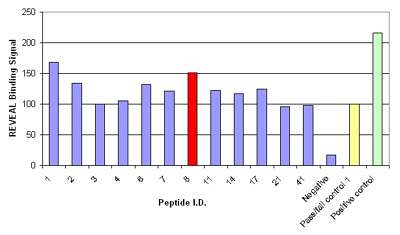
Figure 1: Results of the MHC-peptide binding assay for controls and the 12 peptides that were considered potential epitopes; binding signal is shown as a percentage relative to the pass/fail control; peptide 8, HPVHAGPIA (red) was further validated by Pentamer staining.
Westrop et al., then carried out Pro5® MHC Class I Pentamer staining, which showed a well-defined Pentamer+/CD8+ population for the B*35:01/HPVHAGPIA epitope (figure 2). Additionally, no naïve or central memory T cells specific for the HA9 peptide were found, indicating a lack of proliferative potential in infected HIV-1+ individuals. Further validation with ELISpot assays showed that the HA9 HPVHAGPIA peptide elicited the highest ex vivo response of the peptides studied.
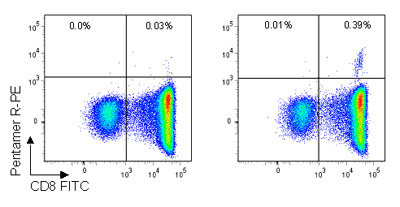
Figure 2: Pro5® Pentamer staining of live lymphocytes gated on CD3+ cells; the left plot shows staining with an allele mismatched negative control Pentamer, the right plot shows staining with the HA9 (B*35:01/HPVHAGPIA) Pentamer. 0.39% of CD3+ live lymphocytes are CD8+/HA Pentamer+ with a background stain of 0.03%.
The author commented that this study “confirms the novel ProImmune technology as a useful and superior tool for revealing potential immunogenic epitopes”, and also highlighted the importance of validating results in parallel using functional assays, such as ELISpot and Pro5® Pentamer staining.
