Your basket is currently empty!
T cell Assays
T cell Assays for Allergenicity Testing
How do Functionally Similar Proteins Compare in their Immunogenicity?
Proteins are an important part of human and animal nutrition yet some proteins, a proportionally very small number, can be allergens and immune system sensitizers. Although many of the food allergens and celiac proteins are well described for their protein sequence and clinical impact there are many challenges, especially in the areas of safety testing and regulation, where a better understanding of the basic biology of allergy sensitization is valuable.
Many times there is no commercial assay platform or other means to generate biologically relevant immunological data for proteins in a standardized and accepted format. Recently, the ProImmune team worked with a company’s internal prospecting group to identify ProImmune as a group with the right capabilities. Having finalized a qualification study design, they chose to utilize T cell assays from ProImmune to investigate the potential to use T-cells as a testing platform for observing a protein’s sensitizing capacity, in vitro.
Many researchers have access to reliable molecular and clinical information on common allergens such as dust mite, and peanut proteins, but the challenge in this customer’s case was to predict whether anmeasuring- unfamiliar protein has the ability to elicit an adverse allergy response in humans. Two qualifying proteins were run through the T cell assay to establish a baseline of quantified immune response to potentially compare other novel proteins against them in the future.
To establish a baseline of experience for a particular functional class of proteins, our customer selected a human enzyme (a self-protein to which humans are expected to be tolerized), and its bacterial orthologue, which is not known to elicit an immune response in humans. They compared the two proteins using CFSE T cell assays. In these assays, an overlapping peptide library is generated from each protein sequence, and the potential for each peptide to elicit CD4+ T cell proliferation in PBMC from a range of donors is tested. Proliferation was measured by CFSE dye dilution.
For this project, we used a panel of 40 healthy donors with HLA haplotypes chosen to match the global population distribution. Each peptide was tested in sextuplicate against each donor. We calculated a cell division index by comparing the proportion of divided (CFSE-dim) CD4+ T cells to the total number of CD4+ T cells, and in turn comparing this figure from stimulated and unstimulated samples (figure 1), and this gives a measure of the likely immunogenicity of each peptide.
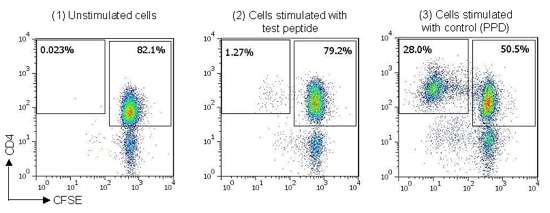
Figure 1: Example staining data from naïve T cell assay, (1) CFSE-labeled T cells cultured in media alone (unstimulated), (2) CFSE-labeled T cells cultured with peptide derived from antigen of interest, (3) CFSE-labeled T cells cultured with control protein (Tuberculin PPD).
As a result of the data presented by ProImmune’s customized report, the customer now has a picture of the performance of two ‘safe’ proteins. The hypothesis that a human protein generates a lower immunogenicity response than a bacterial protein with the same function appears to be borne out by the data. The next step is to consider comparing other proteins against these established “standards” (and to the assay positive controls). The customer noted that the advantage of ProImmune’s approach is the absence of a need for full-length purified protein, allowing us to screen many candidate proteins relatively cheaply. The workflow now established with ProImmune will allow our customer to supply regulatory agencies with a comprehensive set of information about their novel proteins.
