Your basket is currently empty!
TheWhoHow
The Who, How and Why of Seafood Allergy
Professor Julio Delgado describes his experiences with ProImmune REVEAL® epitope discovery
 |
I’m based at the University of Utah School of Medicine, where I hold an Assistant Professorship and spend part of my time working on the characterization of peptide epitopes. It’s an exciting field- the understanding of epitopes and their MHC restriction is crucial in clinical immunology and in many disease states. As a team we’ve developed and standardized mass-spectrometric techniques for the sequencing of peptides eluted from MHC class I and class II molecules. Using this approach we are characterizing MHC peptide epitopes involved in autoimmune and allergic diseases, and T-cell responses to infectious agents. One recent project of ours has been identifying the epitopes involved in triggering allergic responses to seafood. |
When ProImmune gave a webcast last year about their REVEAL® antigen characterization platform, I was immediately intrigued by the possibility of working with them, and saving ourselves months of research in the lab. I decided to try out ProImmune’s capabilities for our seafood allergy study – we knew there must be epitopes for one particular protein but we hadn’t had time to fully characterize them. We analyzed binding to 9 HLA-DR alleles and 4 HLA-DQ alleles. We chose the alleles based on patient HLA haplotypes and since ProImmune offer so many different alleles for analysis it was no problem for them to accommodate our needs. The advantage of ProImmune’s in vitro system is that we could characterize binding to each HLA molecule individually and not have to guess which of the range of HLA molecules expressed by our patients was responsible for antigen presentation.
We used an overlapping library of peptides from our protein, and also spiked in a couple of controls alongside ProImmune’s own control antigens. ProImmune investigated the binding and stability of our peptides with our chosen HLA types, and drew the data together for us.
|
|
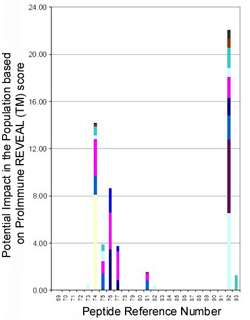
Figure 1: Graph shows potential impact in the population based on ProImmune REVEAL®binding assay scores, for peptides from a protein implicated in seafood allergy responses. The results shown are for peptides 69-93, 14 of the 93 peptides from Prof. Delgado’s project. Each colour represents a different HLA allele, and the bar height indicates potential impact in the global population. Clusters of hit peptides indicate an epitope, for example peptides 74-77 show strong binding across several alleles. Peptide 92 was a positive control chosen by Prof. Delgado. |
Their pan-allele analysis of our data (Figure 1) gave us a picture of the likely impact of each peptide epitope in the global population, and showed a couple of clustered hits across several alleles, which could well be real epitopes.
We’ve been validating the hits using ELISpot performed on blood from our patient cohort. Further characterization of antigenic binding to HLA class II molecules in terms of peptide stability and cytokine production is providing us with critical insights about the immunological mechanisms of antigen presentation that selectively evoke CD4 TH2 cell responses in patients with seafood allergy.
Through our research we hope to facilitate the development of epitope-based tolerizing therapies and immunodiagnostic tools in patients with seafood allergy across different populations.
This work was carried out at the University of Utah in Julio Delgado’s Laboratory.