Your basket is currently empty!
MutaMap™ Mutational Activity Map
MutaMap® Mutational Activity Map
high throughput service for mutational activity maps of therapeutic proteins
A new key tool for developability engineering
When it’s time to make final engineering decisions for your antibody or protein, MutaMap® can help evaluate which individual point mutations to pursue. MutaMap® is an in vitro assay system that helps explore the effect of substituting each amino acid in at each position in a protein sequence one by one with all 19 possible substitutions and find out the effect on protein activity. For example a sequence stretch of 100 amino acids will result in up to 2000 mutants to explore.
|
The approach of MutaMap® is simple. Each position of a protein of interest is mutated by site directed mutagenesis, expressed and tested for its affinity/activity. MutaMap® does not use any surrogate measurement for affinity or activity. Cell free in vitro translation of proteins is combined with solution titration assays to measure affinity/activity. Both methods are optimized for high throughput processing of samples while still allowing for accurate measurements of affinity/activity. The technology is particularly suitable for investing the ligand binding interactions of high affinity monoclonal antibodies, down into the high femtomolar range where other approaches such as SPR struggle to deliver high throughput results. |
|
Mutations investigated in CDR1 and CDR3 of Avastin® heavy chain variable domain
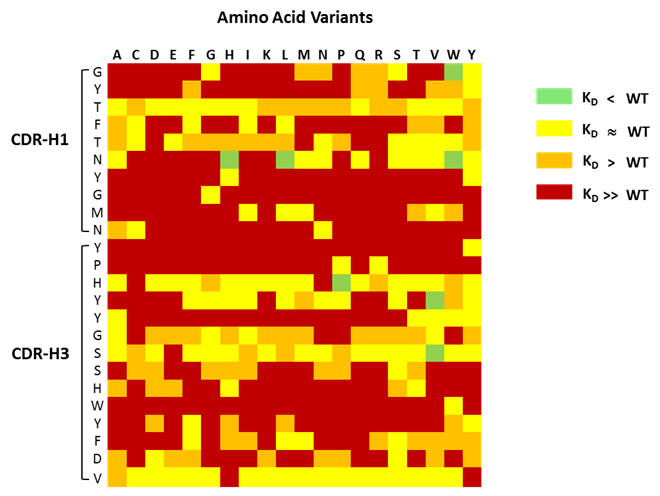
Figure 1: Shown above is an example of a mutagenesis heatmap generated for CDR1 and CDR3 from Avastin® heavy chain variable fragment.
For binding pair interactions MutaMap® uses high throughput solution equilibrium titration (SET) immunoassays to determine the binding affinity for each construct tested.
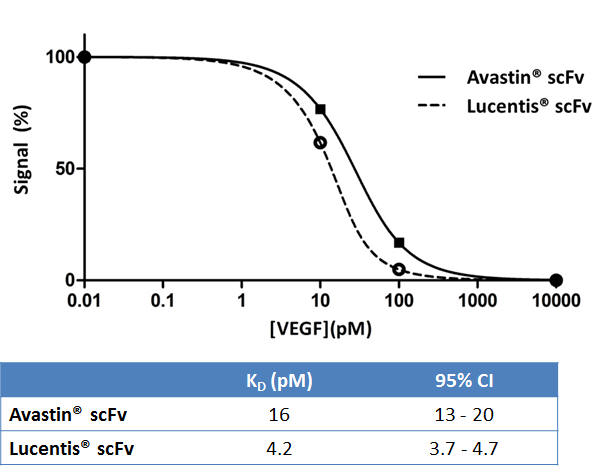
Figure 2: SET results for measuring the affinity of Avastin® scFv and Lucentis® scFv. The resulting titration curve is regressed according to the relevant mass action binding laws. The robustness of the SET approach ensures that this high throughput assay works well for affinities in the single digit picomolar range, as demonstrated by the tight confidence intervals.
What does MutaMap® show you?
MutaMap® delivers a heat map for your protein (see Figure 1 above) that shows you which point mutations lead to an increase, decrease, or no change in affinity (or other activity) or non-function of the protein when interacting with one or more of its binding partners. Effectively you can learn which mutations, one by one, are likely to be permissible or favourable in your protein in terms of the key property of binding to a ligand.
MutaMap® therefore allows you to make informed protein engineering decisions for a range of key developability objectives which include:
- Improving activity/affinity by cherry picking mutations, including in the CDRs of mAbs
- De-immunization while maintaining or even improving protein activity
- Altering cross reactivity, e.g. for improved cross-species reactivity
- Improved humanization or provision of other engineered features
- Stability/Manufacturability and other key developability improvements, achievable through deliberate point mutations
- Prolonging half-life
- Developing unique new composition of matter IP
Focus on position S105 in CDR-H3 of Avastin® heavy chain variable domain

Figure 3: What is clear is that the MutaMap® heatmap shows permissible mutations, especially in CDR-H3 in cases that are not normally considered conservative, e.g. in position S105. This opens up choices for re-designing the molecule that would not normally be available based on computational assessments.
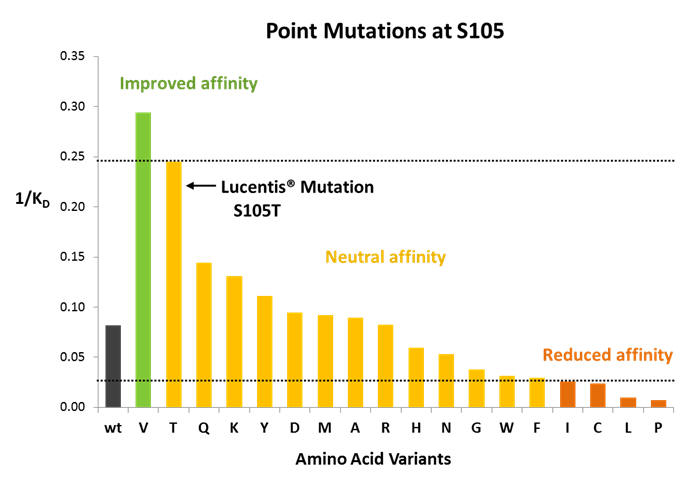
Figure 4: Position S105 in Avastin® CDR-H3 is mutated to T in in Lucentis®. MutaMap® reconfirms that this mutation is indeed beneficial for improving binding. It also shows that a number of other mutations are available to match or improve the affinity of the construct over the Avastin® wild type.
How does MutaMap® compare to molecular evolution technologies?
Molecular evolution techniques such as phage display and other phenotype-genotype coupled randomization techniques are most commonly used in the affinity maturation process for monoclonal antibodies and other binding scaffolds. The advantage of these technologies is that they help explore a very large sequence space of combined mutations.
There comes a point however when final decisions have to be made on the implementation of a protein sequence where individual point mutations may be considered in an antibody or therapeutic protein to meet a variety of design objectives. Randomized molecular evolution is not appropriate for this step. Exploring individual point mutations is nothing new, but it has been difficult to carry this step out in very high throughput way, especially where the objective is to clone and express every mutant and then measure its affinity/activity with reasonable accuracy. This is what MutaMap™ can achieve.
Where does MutaMap® fit in as part a project for e.g. generating a candidate monoclonal antibody for clinical development?
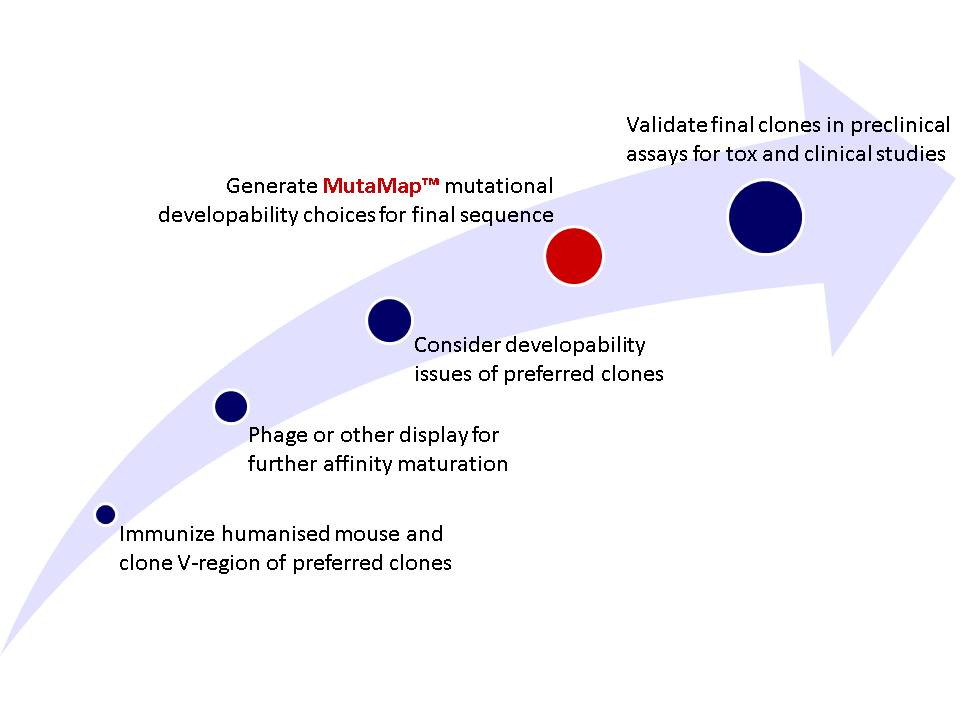
Figure 5: Example work flow for pre-clinial protein engineering of therapeutic monoclonal antibody; individual projects may differ.
How long does a MutaMap® project take?
Our objective is to complete medium size projects of exploring 500-2000 mutations in approximately 8-12 weeks from receiving the customer’s protein sequence. Larger projects will take slightly longer, depending on complexity.
What is the stepwise process for carrying out MutaMap®?
- First we will discuss with you the sequence space you want to explore. This may be the known paratope of a protein or the CDR and flanking regions of a monoclonal antibody.
- To save time and cost you may not want to explore substituting amino acids into the sequence that are considered highly non-conservative or prone to degradation when exposed. We will also discuss what you know about the binding interaction of your protein with its target, whether you have working immunoassays for this interaction and whether the wild type protein and isolated ligand is available to work with in an immunoassay system. Depending on the nature of the interaction and the binding partners we will ensure that the base assay of binding wild type protein to target works well.
- Once these details are agreed and the project is initiated we will run the mutagenesis agreed for each position, express the protein at small scale and carry out the equilibrium binding titration affinity measurement in high throughput.
What will you get?
A final technical report delivered via our secure webserver showing you the affinity or activity determination for the wild type and each mutant with confidence interval. This will be presented in various formats for ease of interpretation, including a standard heatmap.
For customers that want to carry out protein antigenicity studies in parallel, these can be carried out in approximately the same timeframe as the MutaMap®. This means that within a period of approximately 8-12 weeks we will have determined experimentally both the putative T cell epitopes and the MutaMap® of permitted mutations in your protein sequence. This information can allow you to proceed with much better informed decisions on how to address immunogenicity related issues for your program while addressing simultaneously other developability related design decisions for your sequence.
